A Proteomic View on Genome-Based Signal Peptide Predictions
- Haike Antelmann1,
- Harold Tjalsma2,
- Birgit Voigt1,
- Steffen Ohlmeier1,
- Sierd Bron2,
- Jan Maarten van Dijl3,4, and
- Michael Hecker1
- 1Institut für Mikrobiologie und Molekularbiologie, Ernst-Moritz-Arndt-Universiät Greifswald, D-17487 Greifswald, Germany; 2Department of Genetics, Groningen Biomolecular Sciences and Biotechnology Institute, 9751 NN Haren, The Netherlands; 3Department of Pharmaceutical Biology, University of Groningen, Groningen, The Netherlands
Abstract
The availability of complete genome sequences has allowed the prediction of all exported proteins of the corresponding organisms with dedicated algorithms. Even though numerous studies report on genome-based predictions of signal peptides and cell retention signals, they lack a proteomic verification. For example, 180 secretory and 114 lipoprotein signal peptides were predicted recently for the Gram-positive eubacterium Bacillus subtilis. In the present studies, proteomic approaches were used to define the extracellular complement of the B. subtilis secretome. Using different growth conditions and a hyper-secreting mutant, ∼200 extracellular proteins were visualized by two-dimensional (2D) gel electrophoresis, of which 82 were identified by mass spectrometry. These include 41 proteins that have a potential signal peptide with a type I signal peptidase (SPase) cleavage site, and lack a retention signal. Strikingly, the remaining 41 proteins were predicted previously to be cell associated because of the apparent absence of a signal peptide (22), or the presence of specific cell retention signals in addition to an export signal (19). To test the importance of the five type I SPases and the unique lipoprotein-specific SPase of B. subtilis, the extracellular proteome of (multiple) SPase mutants was analyzed. Surprisingly, only the processing of the polytopic membrane protein YfnI was strongly inhibited in Spase I mutants, showing for the first time that a native eubacterial membrane protein is a genuine Spase I substrate. Furthermore, a mutation affecting lipoprotein modification and processing resulted in the shedding of at least 23 (lipo-)proteins into the medium. In conclusion, our observations show that genome-based predictions reflect the actual composition of the extracellular proteome for ∼50%. Major problems are currently encountered with the prediction of extracellular proteins lacking signal peptides (including cytoplasmic proteins) and lipoproteins.
As soil microorganisms, Bacillus species secrete numerous enzymes, enabling them to degrade a variety of substrates and survive in a complex and continuously changing environment. The best-studied representative of this genus isBacillus subtilis. The genome sequence of this organism was published in 1997, representing the first complete genome sequence of a Gram-positive eubacterium (Kunst et al. 1997). Since then, muchBacillus-related research has been focused on the functional analysis of the identified genes and their expression (see the Micado and JAFAN databases,http://Locus.jouy.inra.fr/cgi-bin/genmic/madbase_home.pl andhttp://bacillus.genome.ad.jp/BSORF-DB.html, respectively ). In contrast, the subcellular localization of the corresponding proteins has received relatively little attention thus far. Very recently, we made a first series of predictions concerning the composition of the so-called secretome of B. subtilis, which, by definition, includes both the secreted proteins and the protein secretion machineries (Tjalsma et al. 2000). In summary, four distinct pathways for protein export from the cytoplasm and ∼300 proteins with the potential to be exported were distinguished. By far, the largest number of exported proteins was predicted to follow the major Sec pathway for protein secretion. In contrast, the recently identifiedtwin-arginine translocation Tat pathway (Jongbloed et al. 2000), a pseudopilin export pathway for competence development, and certain ATP-bindingcassette (ABC) transporters can be regarded as special-purpose pathways, through which only few proteins appear to be transported (Tjalsma et al. 2000).
Compared with eukaryotes such as yeast (Drawid and Gerstein 2000;Drawid et al. 2000), vegetative cells of B. subtilis have a limited number of subcellular compartments, the cytoplasm being confined by the cytoplasmic membrane, which, in turn, is surrounded by the cell wall. Notably, the cell wall is a thick (10–50 nm) extracytoplasmic compartment composed of peptidoglycan and anionic polymers, such as teichoic and teichuronic acids (Archibald et al. 1993). Even though the cell wall of B. subtilis may be functionally equivalent to the Gram-negative periplasm (Pooley and Karamata 1996), proteins that are active in this compartment have to be retained with specific cell-wall-binding domains. In fact, ∼56% of the proteins containing a potential amino-terminal signal peptide for export from the cytoplasm are predicted to be secreted into the growth medium, because these proteins lack specific membrane or cell-wall-retention signals (Tjalsma et al. 2000).
During or shortly after the translocation of (pre-)proteins across the membrane, their amino-terminal signal peptide is removed by SPases (signal peptidases). In B. subtilis, two major classes of signal peptides can be distinguished on the basis of the SPase recognition sequence (Tjalsma et al. 2000). The first class is composed of 180 typical signal peptides, which are present in secretory pre-proteins that are cleaved by one of the five type-I SPases (SipS-W) of B. subtilis (Tjalsma et al. 1997,1998). The second major class of signal peptides is present in 114 (putative) pre-lipoproteins, which are cleaved by the lipoprotein-specific (type II) SPase of B. subtilis (Lsp;Tjalsma et al. 1999). The most important difference between signal peptides of lipoproteins and secretory proteins is the presence of a conserved lipobox in lipoprotein precursors. This lipobox contains an invariable cysteine residue that is lipid modified by the lipoprotein diacylglyceryl transferase (Lgt), prior to precursor cleavage by SPase II (Leskelä et al. 1999; Tjalsma et al. 1999). The exported lipid-modified proteins remain anchored to the membrane by their amino-terminal lipid-modified cysteine residue, which serves as a membrane retention signal. Notably, 13 proteins appear to contain a potential cell-wall-retention signal in addition to an amino-terminal signal peptide (for review, see Tjalsma et al. 2000).
The fact that exported B. subtilis proteins are not retained by an outer membrane, as encounterd in Gram-negative eubacteria, makes this Gram-positive eubacterium a preferred organism for the proteomic verification of genome-based signal peptide predictions. In the present studies, we have accomplished this by two-dimensional (2D) gel electrophoresis and subsequent mass spectrometry of extracellular proteins. The results show that our previous predictions of signal peptides with an SPase I cleavage site are largely correct. However, a significant number of membrane proteins, predicted lipoproteins, and even proteins without a signal peptide was also identified in the growth medium. These observations imply that, in addition to the known mechanisms for protein export, B. subtilis can avail itself of alternative mechanisms to release and/or maintain such proteins into the external environment.
RESULTS
The Extracellular Proteome of B. subtilis 168
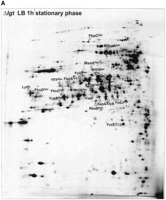
The highest levels of protein secretion are usually observed when cells of B. subtilis are grown in rich medium, in particular during the stationary growth phase. Therefore, to create a first inventory of the extracellular proteome, we have used B. subtilis cells grown in L-broth. Samples were taken during exponential growth and after entry into the stationary phase. Next, extracellular proteins secreted into the growth medium were precipitated and subjected to 2D gel electrophoresis. As shown in Figure 1, most of the extracellular proteins, which are only weakly detectable during exponential growth, are induced upon entry into the stationary phase. Of the ∼200 visible extracellular proteins, 75 different proteins could be identified by matrix assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry as marked in the 2D master gel for the extracellular proteome (Fig. 1B). Seven additional extracellular proteins were identified in the medium of phosphate starvation-induced cells (Antelmann et al. 2000).
The extracellular proteome of B. subtilis 168. Cells ofB. subtilis 168 were grown in L-broth and proteins in the growth medium were harvested during exponential growth (OD540 = 0.4) (A) and 1 h after entry into the stationary phase (B; master gel for the extracellular proteome). After precipitation with TCA, the extracellular proteins were separated by 2D PAGE as described in the Methods section. The proteins identified by mass spectrometry are indicated on the gel and listed in Table 1. These gels are available in the Sub2D proteome database (http://microbio2.biologie.uni-greifswald.de:8880/). (See following page for panel B.)
The 82 identified extracellular proteins include 50 proteins to which a function had been assigned previously, and 32 Y proteins of apparently unknown function (Table 1A,B). Nevertheless, a possible function could be attributed to 19 Y proteins on the basis of their amino acid sequence similarity to proteins with a known function. Taken together, it seems that the identified extracellular proteins of B. subtilis 168 include 18 enzymes related to the metabolism of carbohydrates, nine proteases or peptidases, two enzymes involved in the metabolism of amino acids, three enzymes involved in the decay of DNA or RNA, one lipase, two alkaline phosphatases, two phosphodiesterases, seven proteins involved in cell-wall biogenesis, eight lipoproteins, of which seven are substrate-binding components of various transport systems (Table 1A, proteins marked lipo), five proteins involved in detoxification, three flagella-related proteins, one putative transcriptional regulator, two proteins involved in protein synthesis and folding (among which is the chaperonin GroEL), five prophage-related proteins, one sporulation-specific protein, and 13 proteins of unknown function. Notably, the relative amounts of 60 identified extracellular proteins are significantly increased during the stationary phase (Table 1C).
Most of the identified extracellular proteins migrate on the 2D gels at a position that is in good agreement with their calculated molecular weight and isoelectric point. It has to be noted that several protein spots are present as pearl chains, which might represent isoelectric focusing artefacts that are due to the trichloroacetic acid (TCA) precipitation during sample preparation. Notably, the high-molecular weight cell-wall-associated proteins WprA and WapA, as well as the bacillopeptidase F (Bpr), the serine protease (Epr), and the major autolysin LytD do not run at, or close to, the estimated position. Most likely, this is due to proteolytic processing upon export from the cytoplasm. In fact, the two major degradation products of WprA, known as CWBP52 and CWBP23, were both detected (Fig. 1B). In addition, several degradation products of WapA could be identified, of which the 58-kD product represented one of the most abundant extracellular proteins. As revealed by amino-terminal sequencing, the latter protein represents the carboxy-terminal end of WapA, starting with Ser1725 (Foster 1993).
Verification of Genome-based Signal Peptide Predictions
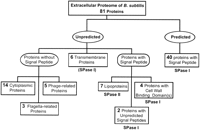
Of the 82 identifed extracellular proteins (Table 1), 48 were previously predicted to contain an amino-terminal signal peptide and no transmembrane segments (Tjalsma et al. 2000): (numbers shown in italics refer to the overview in Fig. 2) 39 with a type I SPase cleavage site, and 9 with a type II SPase cleavage site. Notably, 4 of the predicted pre-proteins with a type I SPase cleavage site also contain typical cell-wall-binding repeats (LytD, WapA, YvcE, and YwtD). In contrast,2 proteins (YweA and YxkC) with a signal peptide and a type I SPase cleavage site were not predicted previously; the signal peptide of YweA was missed by the SignalP algorithm for unknown reasons, and the signal peptide of YxkC because it is unusually long (Table 1A). As shown by amino-terminal sequencing, one of the 9 proteins with a predicted SPase II cleavage site and no potential transmembrane segments, YrpD, was processed at a typical cleavage site for SPase I (Table 1A), suggesting that this protein is not a lipoprotein. Moreover, the potential SPase II site of YrpD (FGTC) was found only once among the 114 predicted lipoproteins of B. subtilis. Similarly, the XynD protein has a potential SPase I cleavage site in addition to a uniquely observed SPase II cleavage site (LLSC) and a transmembrane segment (Tjalsma et al. 2000). In the following report, YrpD and XynD are, therefore, no longer referred to as lipoproteins.
Overview of identified and predicted extracellular proteins of B. subtilis 168. Extracellular proteins, as listed in Table 1, were classified as predicted or unpredicted with respect to their extracellular localization on the basis of previous genome-based predictions of signal peptides and retention signals (Tjalsma et al. 2000). Proteins with signal peptides are further classified on the basis of their cleavage sites for SPase I or II, and the presence of cell-wall-binding domains. Note that potential pre-proteins with an SPase I cleavage site and predicted transmembrane segments were excluded from previous predictions. XynD, which has a predicted SPase II cleavage site in addition to a potential transmembrane segment, is counted as a transmembrane protein; both predictions are, however, probably wrong (see Results section and Table 1A). The indicated numbers correspond to numbers printed in italics in the Results section.
The extracellular proteins that were not included previously in our listings of proteins with a cleavable signal peptide (Tjalsma et al. 2000) form a heterogeneous group as follows: 6 have potential transmembrane segments according to the TopPred algorithm of Sipos and von Heijne (1993) (Ggt, XynD, YfkN, YhcR, PbpA, and YfnI), and 22 other proteins lack an amino-terminal signal peptide of known type (Table 1). Upon reinvestigation with the TMHMM algorithm for the prediction of transmembrane segments (for review, see Nilsson et al. 2000), the presence of transmembrane segments in 2 of the 6 extracellular proteins (Ggt and XynD; Table 1A) was not confirmed. Finally, the group of extracellular proteins without a typical signal peptide includes3 flagella-related proteins (Hag, FlgK, and FliD), 5prophage-related proteins (XepA, XkdG, XkdK, XkdM, and XlyA), and14 hydrophilic proteins (Table 1B; Fig. 2). The latter proteins were identified as abundant cytoplasmic proteins (H. Antelmann and M. Hecker, unpubl.).
The Extracellular Proteome of the Hyper-secretingdegU32(hy) Mutant
The induction of extracellular degradative enzymes of B. subtilis at the end of the exponential growth phase is dependent on the DegSU two-component system (Lepesant et al. 1972; Ayusaw et al. 1975). Certain mutations in the degS or degU genes, causing more stable phosphorylation of DegU, result in the hyperproduction of degradative enzymes (Hy-phenotype; Henner et al. 1988; Kunst et al. 1988). Despite the fact that numerous studies on the DegSU system have been documented, a comprehensive description of all DegSU-regulated extracellular proteins is still lacking. For this reason, we analyzed the extracellular proteome of the hyper-secretingdegU32(hy) strain, which exhibited dramatic changes compared with the parental strain. All identified extracellular proteins that are up- or down-regulated in this hyperphosphorylateddegU32(hy) mutant are listed in Table2. As expected, the alkaline protease AprE represented the most abundant protease secreted into the medium. In addition, six other proteases/peptidases, including the bacillopeptidase Bpr, the serine protease Vpr, the metalloproteases Mpr and NprE, the γ-glutamyltranspeptidase Ggt, and the aminopeptidase YwaD were overproduced in the degU32(hy) mutant strain. Of the secreted enzymes involved in the metabolism of carbohydrates, the α-amylase AmyE, β-glucanase BglS, xylanase YnfF, and pectate lyase PelB were found to be present at significantly elevated levels. Finally, increased amounts of two enzymes involved in the degradation of nucleotides, the ribonuclease YurI and 2‘,3′-cyclic-nucleotide 2‘-phosphodiesterase YfkN, were detected in the medium of thedegU32(hy) strain, indicating that the corresponding genes are new members of the DegSU-regulon.
Extracellular Proteins of Bacillus subtilis 168 with Typical Export Signals1
Proteins of Bacillus subtilis 168 Present in the Medium Without Typical Export Signals1
Extracellular Proteins Present at Higher Levels During the Stationary Phase
Extracellular Proteins Probably under DegU Control
Notably, the degU32(hy) mutation is known to display pleiotropic effects. Thus, unlike the parental strain 168, thedegU32(hy) mutant does not only show an increased secretion of degradative enzymes, but it can also sporulate in the presence of glucose, displays a filamentous morphology, lacks flagella, and is unable to develop competence for DNA binding and uptake (Kunst et al. 1974). Consistent with some of these phenotypes, the autolysins LytD and YwtD, flagellin Hag, flagellar hook-associated proteins FlgK and FliD, and the wall-associated protein WapA (the genes of which are all regulated by the alternative ς factor ςD for motility and chemotaxis; Helmann et al. 1988) were absent from the medium of thedegU32(hy) mutant (Fig. 3). Furthermore, the relative amounts of two extracellular proteins of unknown function, YlqB and YxkC, were strongly reduced.
The extracellular proteome of B. subtilis degU32(hy). Cells ofB. subtilis degU32(hy) were grown in L-broth and proteins in the growth medium were harvested 1 h after entry into the stationary phase. After precipitation with TCA, the extracellular proteins were separated by 2D PAGE as described in the Methods section. The proteins identified by mass spectrometry are indicated. Extracellular proteins of which the synthesis is induced in the degU32(hy) mutant are boxed, and those of which the synthesis is reduced are printed in bold (see also the listing in Table 2). This gel is available in the Sub2D proteome database (http://microbio2.biologie.uni-greifswald.de:8880/).
The Membrane Protein YfnI is Processed by the Signal Peptidases SipT and SipV
Previous studies on the five type I SPases of B. subtilishave shown that these enzymes have different, but overlapping substrate specificities, as exemplified with the α-amylase AmyQ (Tjalsma et al. 1997, 1998). To further investigate possible differences in the substrate specificities of the type I SPases of B. subtilis, we analyzed the extracellular proteomes of single SPase mutants lackingsipS, sipT, sipU, sipV, orsipW. Surprisingly, no major differences in the extracellular protein patterns of these mutant strains were observed (data not shown). Therefore, we also analyzed the extracellular proteomes of double, triple, and quadruple sip mutant strains as listed in Table 1. Strikingly, all 2D gels with extracellular proteins of multiple sip mutant strains displayed nearly identical protein patterns with only one exception, the mutant strains lacking bothsipT and sipV showed strongly reduced extracellular levels of the membrane protein YfnI (Fig.4). This result implies that SipT and SipV are responsible for the specific cleavage of YfnI.
Processing of the membrane protein YfnI by the type I signal peptidases SipT and SipV. Single and multiple signal peptidase (sip) mutants of B. subtilis were grown in L-broth, and proteins in the growth medium were harvested 1 h after entry into the stationary phase. After precipitation with TCA, the extracellular proteins were separated by 2D PAGE as described in the Methods section. The three spots that correspond to the YfnI protein are encircled.
Reduced Levels of Non-lipoproteins in the Medium of an SPase II Mutant
The lipoprotein-specific SPase II of B. subtilis has been shown previously to be indirectly required for the folding and secretion of the α-amylase AmyQ, a non-lipoprotein (Tjalsma et al. 1999). To investigate to what extent other secreted proteins are affected by the absence of SPase II, the extracellular proteome of anlspA mutant strain was analyzed. Interestingly, protein secretion was significantly impaired by the lspA mutation, as evidenced by the fact that the protein content of the culture supernatant was reduced to ∼25% of that of the parental strain 168. More specifically, two abundant extracellular proteins of the parental strain, the α-amylase AmyE and the protein of unknown function YolA, were completely absent from the extracellular proteome of the lspA mutant (compare Fig. 1B and Fig.5). Furthermore, the relative amounts of a variety of other extracellular proteins were strongly reduced as exemplified by the chitosanase Csn, serine proteases Epr, lipase LipA, glycerophosphoryl diester phosphodiesterase GlpQ, major autolysin LytD, penicillinase PenP, phage-related proteins XepA, XkdK, XkdM, and XlyA, and certain proteins of unknown function (YncM, YrpD, YwoF, YxaK, and YxkC; Table 3). Unexpectedly, we observed a significant increase in the level of two processed forms of the wall protease WprA (CWBP23 and CWBP25) in the medium of the lspAmutant strain. Similarly, the extracellular levels of two typical lipoproteins, MntA and YxeB, were strongly increased, showing that these proteins were not effectively retained in the membrane of thelspA mutant. In contrast, the extracellular levels of the lipoproteins OppA and YclQ were not affected by the lspAmutation (Fig. 5). Taken together, these findings show that the absence of SPase II has pleiotropic effects on the composition of the extracellular proteome.
The extracellular proteome of SPase II mutant cells of B. subtilis. Cells of B. subtilis ΔlspA, which lack SPase II, were grown in L-broth and the proteins in the growth medium were harvested 1 h after entry into the stationary phase. After precipitation with TCA, the extracellular proteins were separated by 2D PAGE as described in the Methods section. The proteins identified by mass spectrometry are indicated. Extracellular proteins that are present at elevated levels are encircled, and extracellular proteins that are present at reduced levels are boxed (see also the listing in Table 3). This gel is available in the Sub2D proteome database (http://microbio2.biologie.uni-greifswald.de:8880/).
Extracellular Proteome Changes in the Absence of SPase II
Shedding of Lipoproteins and Autolysins by a Diacyl-glyceryl Transferase Mutant
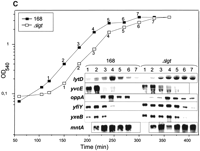
As indicated above, eight potential lipoproteins were shown to be present on the extracellular proteome of the parental strain, four in L-broth (MntA, OppA, YclQ, and YfmC), and four in minimal medium in response to phosphate starvation (PstS, YcdH, YdhF, and YqiX; Table 1A). In addition, elevated levels of MntA and YxeB were found on the extracellular proteome of the lspA mutant strains. To further explore the factors required for lipoprotein processing and retention in the cell, the composition of the extracellular proteome of anlgt mutant, which is defective in the lipid-modification of lipoproteins (Leskelä et al. 1999), was analyzed. Strikingly, the extracellular protein pattern of the lgt mutant grown in L-broth was completely different from that of the parental strain, exhibiting ∼35 additional spots that were not, or only very weakly, present in the parental pattern (compare Fig. 1B and Fig. 6A; Table4). Furthermore, the extracellular levels of the predicted lipoproteins OppA and MntA, and several autolytic enzymes were significantly increased by the lgt mutation (Fig. 6A). The latter comprise YvcE, a predicted cell-wall-binding protein of the endopeptidase II family, the major N-acetylglucosaminidase LytD, and the host cell lysis enzymes XepA and XlyA encoded by the prophage PBSX. Moreover, the extracellular level of YwtF, a potential gene regulator similar to LytR, which is involved in the regulation of autolysins, was significantly increased (Fig. 6A). By use of MALDI-TOF mass spectrometry, nine of the additional extracellular proteins appearing in the medium of the lgt mutant were identified. Notably, all of these are predicted lipoproteins (Table 4), including binding proteins for the uptake of iron or ferrichrome (FeuA, FhuD, YfiY, and YxeB), multiple sugars (MsmE), and ribose (RbsB), as well as two proteins involved in cell-wall metabolism, the penicillin-binding protein PbpC and the putative D-alanyl-D-alanine carboxypeptidase YodJ. These observations show that cells lacking the diacyl-glyceryl transferase shed lipoproteins and autolysins into their growth medium.
The extracellular proteome of diacylglyceryl transferase mutant cells of B. subtilis. (A) Cells of B. subtilisΔlgt, which lack the diacylglyceryl transferase, were grown in L-broth and proteins in the growth medium were harvested 1 h after entry into the stationary phase. After precipitation with TCA, the extracellular proteins were separated by 2D PAGE as described in the Methods section. The proteins identified by mass spectrometry are indicated. Non-lipoproteins that are present at elevated levels in the medium of the Δlgt strain are printed in bold; lipoproteins that are released into the medium of the Δlgt strain are printed in bold and marked with lipo (see also the listing in Table 4). This gel is available in the Sub2D proteome database (http://microbio2.biologie.uni-greifswald.de:8880/). (B). Comparison of extracellular and cell-associated lipoproteins of phosphate-starved lgt mutant and parental (168) cells. Phosphate starvation was provoked by growing the cells in a minimal medium with limiting amounts of phosphate as described previously (Antelmann et al. 2000). Cellular proteins and proteins in the growth medium were harvested 1 h after entry into the stationary phase provoked by phosphate starvation, and samples for 2D PAGE were prepared as described in the Methods section. The identity of all spots in the PstS cluster was confirmed by mass spectrometry. Lipoproteins that are present at increased levels in the medium due to the lgtmutation are marked with lipo and listed in Table 4. (C) Transcript analyses of the lytD, yvcE,oppA, yfiY, yxeB, andmntA genes ofB. subtilis Δlgt and the parental strain 168. Total RNA was isolated from cells grown in L-broth at different time points (1–7), as indicated in the growth curves. For the Northern blotting experiments, samples of 10 μg of total RNA, containing nearly identical amounts of rRNA (internal standard), were applied in each lane.
Shedding of Lipoproteins and Autolysins by Bacillus subtilis Δlgt
Upon phosphate starvation, the lipoproteins PstS (a phosphate-binding protein) and YdhF are strongly induced and released into the growth medium of B. subtilis 168 (for review, see Antelmann et al. 2000). This raised the question as to whether increased amounts of these two proteins would be shed into the medium of the phosphate-starved lgt mutant strain. As shown in Figure 6B, this was the case. In addition, the extracellular levels of the predicted lipoproteins OppA, PbpC, YfiY, YusA, and YxeB, which were also identified when the lgt mutant was grown in L-broth, were significantly increased (Fig. 6B, data not shown). Finally, the amounts of four newly identified lipoproteins present in minimal medium were increased by the lgt mutation. The latter proteins include binding proteins for the uptake of glycine-betaine (OpuAC), zinc (YcdH), amino acids (YqiX), and a lipoprotein of unknown function (YrpE). As shown previously, PstS is one of the major cellular (note that membrane-attached translocated lipoproteins fractionate with cells) phosphate starvation-induced proteins (Eymann et al. 1996), we investigated the electrophoretic properties of cell-associated PstS in phosphate-starved lgt mutant cells. Figure 6B shows that PstS was shifted to a higher Mw and pI compared with the parental strain, suggesting that these cell-associated forms of PstS might represent non-modified lipoprotein precursors. Unfortunately, the amino-termini of these PstS precursors were blocked for sequencing, most likely due to the presence of an amino-terminal formyl-methionine residue.
To determine whether the shedding of lipoproteins and autolysins in thelgt mutant is accompanied by increased transcription of the corresponding genes, Northern hybridization analyses were performed using specific RNA probes. Thus, the transcription levels of four lipoprotein-encoding genes (mntA, oppA,yxeB, and yfiY) and four autolysin-encoding genes (lytD, yvcE, xlyA, and xepA) in thelgt mutant strain were compared with the respective transcription levels in the parental strain in a growth phase-dependent manner. The results showed that the mRNA levels specific foroppA, mntA, yvcE (Fig. 6C), xepA, and xlyA (data not shown) were not significantly influenced by the lgt mutation. In contrast, the lgt mutant produced increased amounts of mRNAs specific for the lytD,yfiY, and yxeB genes during the stationary phase (Fig. 6C). Taken together, these results indicate that there is no causal relationship between the shedding of lipoproteins and autolysins into the growth medium of an lgt mutant strain and the level of transcription of the corresponding genes.
Protein Shedding by the lgt Mutant Due to Limited Retention and Proteolytic Shaving
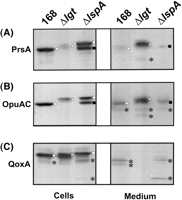
It was shown previously that the lipoprotein PrsA was released as an unmodified precursor into the medium of a lgt mutant strain ofB. subtilis (Leskelä et al. 1999). To investigate the mechanism of pre-lipoprotein release by the lgt mutant, the amino-termini of six released lipoproteins were determined. The results showed that the released lipoproteins MntA, OppA, YclQ, YfiY, YfmC, and YxeB were all cleaved one residue carboxy-terminally of the predicted site for processing by SPase II, resulting in the loss of the amino-terminal cysteine residue of the membrane-anchored lipoprotein (data not shown). Notably, the same alternative processing was observed for the OppA and YclQ proteins that were present in the medium of the parental strain (data not shown). In addition, the shedding of OpuAC was investigated by Western blotting, by use of the lipoprotein QoxA (which has two transmembrane segments) as a negative control, and PrsA as a positive control. Furthermore, the lspA mutant strain was used to visualize lipid-modified precursor forms of OpuAC, PrsA, and QoxA. As shown in Figure 7, pre-OpuAC was released into the medium of the lgt mutant, like pre-PrsA. In contrast, no pre-QoxA was present in the medium of the lgtmutant strain, showing that the release of pre-OpuAC and pre-PrsA is not due to cell lysis.
Shedding of lipoprotein precursors by diacylglyceryl transferase mutant cells of B. subtilis. To investigate in which form the lipoprotein OpuAC is released into the growth medium of lgtmutant cells, Western blotting experiments were performed with cellular and growth medium fractions of B. subtilis Δlgt, and the control strains 168 and Δlsp. Furthermore, the PrsA and QoxA proteins were used as control lipoproteins. Cells were grown in L-broth at 37°C. Approximately 1 h after entry into the stationary phase, cells were separated from the growth medium by centrifugation. Next, extracellular proteins were concentrated by TCA precipitation. Equivalent amounts of cell and medium fractions were used for SDS-PAGE, Western blotting and immunodetection with antibodies specific for PrsA (A), OpuAC (B), or QoxA (C). Unmodified lipoprotein precursors (white triangles), lipid-modified lipoprotein precursors (white squares), mature lipoproteins (white circles), alternatively processed mature-like lipoproteins (black squares), and degradation products of lipoproteins (black stars) are indicated.
Finally, as indicated above, the autolysin LytD (95.4 kD) was detectable in the extracellular proteome of the parental strain in the form of two proteins of ∼45 and 37 kD (Fig. 1B). Compared with the parental strain, the abundance of the 37-kD product of LytD was significantly increased in the extracellular proteome of thelgt mutant (Fig. 6A). Similarly, the YvcE protein (50.8 kD) was present in the extracellular proteome of the lgt mutant in the form of two proteins of ∼25 and 30 kD, respectively. These observations imply that LytD and YvcE are subject to proteolysis, in particular in the lgt mutant. In conclusion, our data show that the shedding of lipoproteins and autolysins into the medium of thelgt mutant is due to proteolysis (shaving) and, in the case of some lipoproteins, inefficient membrane retention (leakage).
DISCUSSION
Secretory proteins of eubacteria are known to perform a variety of very important remote control functions for their survival in the environment, such as the provision of nutrients, cell-to-cell communication, detoxification of the environment, or the killing of potential competitors. More specifically, the extracellular proteins of pathogenic eubacteria seem to play critical roles in virulence. For these reasons, various studies have been initiated lately to define the extracellular proteomes of pathogens (for review, see Jungblut et al. 1999; Rosenkrands et al. 2000; Lei et al. 2001; Ziebandt et al. 2001). Strikingly, the extracellular proteomes of bacilli, which are among the organisms known to secrete the largest amounts of proteins, have remained largely unexplored thus far. In our present studies that build upon the recent identification of 23 extracellular proteins of B. subtilis 168 (Hirose et al. 2000), we have defined 62 additional extracellular proteins of this Gram-positive eubacterium. In addition, we have monitored, for the first time, how the extracellular proteome of B. subtillis changes in response to mutations that are known to affect the synthesis and/or processing of exported pre-proteins. Most importantly, the present data have allowed a proteomic verification of our recent genome-wide prediction of protein export signals of B. subtilis. Even though the process of protein secretion by B. subtilis has been documented fairly well (for review, see Tjalsma et al. 2000), various results were completely unexpected. These include the export of cytoplasmic proteins, processing of a native membrane protein by type I SPases, and the release of 11 normally cell-associated lipoproteins into the growth medium by a mutant affected in pre-lipoprotein modification and processing.
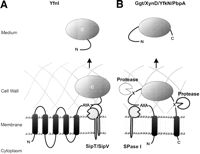
A total number of 50 identified extracellular proteins has a typical signal peptide with a cleavage site for SPase I. In addition, the membrane protein YfnI is cleaved by type I SPases, at an unexpected site (see Table 1). It is interesting to note that, in contrast to our previous predictions (85%), all of these cleavage sites contain an alanine residue at the −1 position relative to the scissile peptide bond (Table 5). Furthermore, the frequency of phenylalanine at the −2 position is significantly lower, whereas that of tyrosine at the −2 position, and alanine and glutamic acid at the +1 position, is significantly higher than predicted previously. Most of these signal peptides are likely to direct the corresponding proteins into the Sec pathway for protein translocation. Only seven contain a potential RR motif, suggesting that they could direct translocation via the Tat pathway. However, Tat-dependent translocation has, thus far, been shown for only one of these seven proteins (PhoD), whereas the RR motif of three other proteins (WapA, WprA, and YfkN) is apparently not recognized by the Tat machinery (Jongbloed et al. 2000). Interestingly, the secretion of none of these proteins was significantly affected by the disruption of single, or even multiplesip genes for type I SPases. This observation confirms the view that the presence of either SipS or SipT is sufficient for efficient precursor processing, and that the type I SPases of B. subtilis have largely overlapping specificities. Surprisingly, the only exception was the SipT/V-dependent cleavage of the membrane protein YnfI. This observation is not only remarkable because YfnI is a polytopic membrane protein, but also because the cleavage site islocated 44 residues carboxy-terminally of the fifth transmembrane segment of this protein (see also Hirose et al. 2000). This suggests that, despite its distant position relative to the transmembrane segment, the SPase I cleavage site of YfnI is accessible to the catalytic sites of SipT and SipV at the extracytoplasmic membrane surface (Fig. 8A). Similarly, the extracytoplasmic domains of three other membrane proteins are most likely liberated from the membrane by proteolysis. In the case of YfkN and YhcR, this seems to be due to amino-terminal processing by SPase I, and carboxy-terminal processing by an unknown protease at the membrane cell-wall interface (Fig. 8B). In contrast, the release of PbpA merely requires amino-terminal processing, which is probably not catalyzed by a known SPase, as PbpA lacks a typical SPase I cleavage site (Fig. 8C).
Amino Acid Residues around (Putative) SPase I Cleavage Sites
Model for the release of proteins with transmembrane segments. (A) Processing of the polytopic membrane protein YfnI by the type I signal peptidases SipT and SipV at the AYA cleavage site. Cleavage occurs carboxy-terminally of A228 (see also Hirose et al. 2000). (B) Cleavage and release of the YfkN and YhcR proteins, which have a predicted amino-terminal signal peptide and a potential carboxy-terminal transmembrane segment. Cleavage is catalyzed by as yet unidentified signal peptidases and/or proteases that are active at the membrane cell-wall interface. (C) Cleavage and release of PbpA, which has a predicted amino-terminal transmembrane segment and lacks a typical SPase cleavage site.
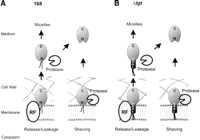
Unexpectedly, eight predicted lipoproteins were identified on the extracellular proteome of B. subtilis 168. As some of these lipoproteins were shown to lack the amino-terminal cysteine residue, they are most likely liberated from the cell by proteolytic shaving after their processing by SPase II (Fig.9A). However, the alternative possibility that, prior to amino-terminal proteolysis, these lipoproteins are released by leakage from the membrane, or even a hypothetical release factor (RF), cannot be excluded presently. For example, active release from the cytoplasmic membrane has been shown for certain lipoproteins of Gram-negative eubacteria (for review, see Yakushi et al. 2000). Unexpectedly, the release of lipoproteins into the medium was not affected in the absence of SPase II and, for unknown reasons, the release of two lipoproteins (MntA and YxeB) was even strongly increased. This implies that the processing defect in the absence of SPase II is bypassed by one or more other proteases that catalyze alternative lipoprotein processing, as shown previously for the lipoprotein PrsA (Tjalsma et al. 1999). Conversely, the secretion of non-lipoproteins was generally reduced in the absence of SPase II. It is tempting to speculate that the latter effect is due to a reduced activity of the essential lipoprotein PrsA, a peptidyl prolylcis-trans isomerase, which is of major importance for extracytoplasmic protein folding (Kontinen and Sarvas 1993; Tjalsma et al. 1999). Alternatively, the reduced extracellular protein levels of the SPase II mutant could, at least in part, be due to the (unexpected) shedding of the catalytic domain CWBP52 of the wall-associated protease WprA into the medium. CWBP52 has been shown to degrade translocated proteins at the membrane cell-wall interface (Stephenson and Harwood 1998; Bolhuis et al. 1999), and it might as well degrade extracellular proteins when shed into the medium in large amounts.
Model for the shedding of lipoproteins. (A) Shedding of the predicted lipoproteins MntA, OppA, PstS, YcdH, YclQ, YdhF, YfmC, and YqiX into the medium of the parental strain is envisaged in at least two ways. First, the lipid-modified mature lipoprotein could be passively (leakage), or actively released from the membrane. Active release would require a dedicated system (the hypothetical release factor RF), as described for lipoproteins of E. coli that are sorted to the outer membrane, or secreted into the growth medium (Yakushi et al. 2000). Thus, although there is no experimental evidence for this presently, lipid-modified mature lipoproteins could be secreted by B. subtilis in the form of micelles. Alternatively, the lipid modification of a released lipoprotein could be lost due to amino-terminal proteolysis. Second, lipid-modified mature lipoproteins could be shaved from the membrane by amino-terminal proteolysis. (B) Lipoprotein shedding by B. subtilisΔlgt can be envisaged to take place in at least two ways. First, unmodified translocated pre-lipoproteins, such as OpuAC and PrsA, could either leak from the membrane, or be actively released into the growth medium by a (hypothetical) release factor (RF). Next, the released pre-lipoproteins could form micel-like structures, or be subject to amino-terminal proteolysis. The latter would result in the presence of mature-like forms in the growth medium as observed for MntA, OppA, YclQ, YfiY, YfmC, and YxeB. Second, these unmodified pre-lipoproteins could be shaved from the membrane by amino-terminal proteolysis.
Lipoprotein shedding was dramatically increased in the absence of the diacylglyceryl transferase Lgt, 11 additional lipoproteins being detectable in the extracellular proteome. Our present observations indicate that there are at least two mechanisms by which this phenomenon can be explained. First, unmodified pre-lipoproteins, such as pre-PrsA (Leskelä et al. 1999) and pre-OpuAC, could be actively or passively released from the membrane (Fig. 9B). Passive release of these pre-proteins might be explained by the fact that the hydrophobic H regions of their signal peptides are too short to span the membrane completely (Table 1A; Tjalsma et al. 2000). Nevertheless, the H region of the signal peptide of PstS appears to be sufficiently hydrophobic to retain the unmodified pre-protein in the membrane. Second, pre-lipoproteins might be released by proteolytic shaving, as evidenced by the fact that at least six extracellular lipoproteins were processed alternatively, lacking the cysteine residue at the +1 position of the mature lipoprotein. However, the latter proteolytic event might as well occur after the release of unmodified pre-lipoproteins from the membrane (Fig. 9B). Proteolytic shaving might also explain the shedding of two autolysins, LytD and YvcE, by thelgt mutant. In contrast, it is presently even less clear how the shedding of the other autolysins, all of which are non-lipoproteins, has to be explained. One possibility is that this phenomenon is caused by changes in the cell wall. These could be due to the malfunction of lipoproteins, which are directly or indirectly involved in cell-wall biogenesis (e.g., DacA, LytA, PbpC, Slp). If so, the function of such lipoproteins would be more strongly affected by the absence of Lgt than the absence of SPase II. Alternatively, lipoprotein malfunction in the lgt mutant might provoke certain undefined stress responses that lead to the shedding of autolysins. The latter idea would be consistent with the observation that the transcription of at least three genes, including the gene for the major autolysin LytD, was increased in the lgt mutant. Thus, induced autolysins, such as LytD, might loosen the peptidoglycan network of the cell wall, thereby causing the shedding of other non-induced autolysins as observed. This view would be supported by the fact that a lytD mutant is affected in protein secretion (Blackman 1998).
The extracellular proteins lacking a typical secretion signal can reach the medium via several routes. First, the five phage-related proteins that were identified have the potential to be secreted via the PBSX prophage-encoded holin XhlB (Longchamp et al. 1994; Krogh et al. 1998), or the SPβ prophage-encoded holins BhlA and BhlB (Regamey and Karamata 1998). Such holins have the potential to oligomerize in the membrane, forming a pore through which the lytic enzymes of bacteriophages, which usually lack a signal peptide, gain access to the cell wall (Young and Bläsi 1992). Second, flagellin and two flagellar hook-associated proteins are, most likely, exported via a dedicated machinery for the assembly of flagella, which is related to the type III secretion machineries of Gram-negative eubacteria (Hueck et al. 1998; Namba et al. 1989). Subsequently, these proteins could be released from the (damaged) flagella. Third, the 13 remaining proteins that lack a signal peptide could be released by (partial) cell lysis, which would be in accord with the relatively high extracellular levels of the major autolysin LytD and the murein hydrolase YwtD. If so, these 13 proteins would be significantly more resistant to degradation by extracellular proteases than the large majority of cytoplasmic proteins. However, the possibility that such proteins escape from the cytoplasm via holins, as recently proposed for Lactococcus lactis (Walker and Klaenhammer 2001), or other (unidentified) protein export pathways, cannot be excluded presently It has to be noted that cytoplasmic proteins, such as aldolase, enolase, elongation factor G, GroEL, and various dehydrogenases, have also been detected in the extracellular proteomes of other eubacteria, such as group A streptococci (for review, see Lei et al. 2000) and mycobacteria (Jungblut et al. 1999; Rosenkrands et al. 2000). Strikingly, ∼60% of the identified extracellular proteins of these organisms lacked a typical signal peptide. By analogy to the fact that at least some of the latter proteins of group A streptococci play an active role in host-pathogen interactions (for review, see Lei et al. 2000), it is conceivable that some of the equivalent extracellular proteins ofB. subtilis support the survival of this organism in its natural habitats, the soil and rhizosphere.
Finally, our proteomic studies provide a first inventory of the numerous degradative enzymes involved in the metabolism of carbohydrates, proteins, lipids, nucleotides, or phosphate that are predominantly secreted during the stationary phase, when the availability of nutrients becomes limiting. These enzymes ensure the cell’s survival during the stationary phase by giving access to alternative nutrient resources. Most importantly, our present observations with the degU32(hy) mutant document for the first time that a subset of 13 degradative enzymes is positively regulated by DegU-phosphate, which starts to accumulate at the end of exponential growth (Ayusaw et al. 1975; Lepesant et al. 1972). Conversely, our results support the view that all members of the ςDregulon, including flagellin, FlgK, FliD, and the major autolysin LytD (Helmann et al. 1988; Mirel and Chamberlin 1989; Kuroda and Sekiguchi 1993; Marquez-Magana and Chamberlin 1994), are negatively regulated by DegU phosphate. In addition, the murein hydrolase YwtD, and two proteins of unknown function (YlqB and YxkC), also seem to be regulated by ςD, as these proteins are absent from the extracellular proteomes of a degU32(hy) mutant, as well as a sigDmutant (H. Antelmann, H. Tjalsma, B. Voigt, S. Ohlmeier, S. Bron, J.M. van Dijl, and M. Hecker, unpubl.).
In conclusion, we have used the power of the high-resolution proteomic technique to get novel insights into the general flow of proteins into the environment of B. subtilis 168. Specifically, our observations shed new light on signal peptide function, the role of signal peptide processing, the importance of signal peptide-independent protein export pathways, the regulation of secretory proteins, and the function of extracellular proteins in general. Consequently, we are convinced that they will serve as major leads for future research on protein transport in B. subtilis and other Gram-positive bacteria that are genetically less amenable than B. subtilis.
METHODS
Plasmids, Bacterial Strains, and Growth Conditions
The bacterial strains and plasmids used are listed in Table6. B. subtilis strains were grown under vigorous agitation at 37°C in L-broth. A phosphate-starvation response was provoked by growing the bacteria in a minimal medium containing 0.16 mM KH2PO4 (Antelmann et al. 2000). Antibiotics were used in the following concentrations: chloramphenicol (Cm), 5 μg/mL; erythromycin (Em), 1 μg/mL; kanamycin (Km), 10 μg/mL; spectinomycin (Sp), 100 μg/mL; tetracycline (Tc), 6 μg/mL; ampicillin (Ap), 50 μg /mL.
Plasmids, Strains, and Primers
DNA Techniques
Procedures for DNA purification, restriction, ligation, agarose gel electrophoresis, and transformation in Escherichia. coli were carried out as described (Sambrook et al. 1989). Enzymes were from Roche Molecular Biochemicals. PCR was carried out with the Pwo DNA polymerase (New England Biolabs) as described (van Dijl et al. 1995).B. subtilis was transformed as described previously (Tjalsma et al. 1998). Single or multiple sip mutations were introduced in B. subtilis 168 by transformation with chromosomal DNA ofB. subtilis 8G5 containing the sipS-Km (ΔS),sipT-Cm (ΔT), sipU-Km (ΔU), sipV-Sp (ΔV), sipW-Tc (ΔW), or lspA-Em (ΔlspA) disruptions (Bolhuis et al. 1996; Tjalsma et al. 1997, 1998, 1999). All chromosomal integrations were verified by PCR.
Analysis of Transcription
Total RNA of B. subtilis was isolated from cells grown in L-broth at different time points during growth using the acidic phenol method of Majumdar et al. (1991). Northern blotting was performed as described previously (Wetzstein et al. 1992). Briefly, internal fragments of the yxeB, yfiY, mntA, oppA, lytD, yvcE, xlyA, andxepA genes were first amplified with primers listed in Table6. Note that the 3′ primer for each of the amplified genes contains a T7 promoter sequence. Next, the amplified fragments were used for the T7 RNA polymerase-directed in vitro synthesis of digoxigenin-labeled gene-specific RNA probes.
Preparation of Cytoplasmic Protein Fractions
Cells grown in phosphate starvation minimal medium were collected by centrifugation 1 h after their entry into the stationary growth phase. Cells were washed twice with 10 mM Tris-HCl, 1 mM EDTA (pH 7.5), and disrupted by sonication (Antelmann et al. 2000). Crude protein extracts were separated from cellular debris by repeated centrifugation for 30 min at 4°C and 15,000 rpm. The protein content of the supernatant fraction was determined according to Bradford (1976) and 80 μg of the crude protein extract was resuspended in 400 μL of immobilized pH gradient (IPG) reswelling solution containing 2 M thiourea, 8 M urea, 1% Nonidet P-40, 20 mM DTT, and 0.5% Pharmalyte (pH 3–10). This sample containing reswelling solution was used for the overnight rehydration of IPG strips in the pH range 3–10. Isoelectric focusing was performed as recommended by the manufacturer (Amersham Pharmacia Biotech). SDS-PAGE in the second dimension was performed as described previously (Antelmann et al. 2000), and the resulting 2D gels were stained with silver nitrate according to Blum et al. (1987). It has to be noted that integral membrane proteins are poorly resolved by the IPG technique due to their hydrophobicity. Consequently, only cytoplasmic proteins and membrane-bound lipoproteins are separated by 2D PAGE.
Preparation of the Extracellular Protein Fraction
B. subtilis cells were grown in 750 mL of L-broth and samples of 250 mL were harvested during exponential growth (OD540 = 0.4), the transition into the stationary phase (t0), or 1 h after the transition into the stationary phase (t1). Cells were removed from the growth medium by centrifugation for 20 min at 4°C and 10,000 rpm. Next, proteins in the medium were precipitated with ice-cold 10% (w/v) trichloroacetic acid (TCA), and collected by centrifugation (40,000g, 45 min, 4°C). The resulting protein pellet was scraped from the wall of the centrifuge tube with a spatula, washed three times with 96% ethanol (v/v) and dried. The extracellular protein fraction of phosphate-starved cells was prepared as described previously (Antelmann et al. 2000). The dried protein pellets were resolved in a solution containing 2 M thiourea and 8 M urea and, subsequently, insoluble material was removed by centrifugation. The protein concentration of the resulting extracellular protein sample was determined according toBradford (1976), and the volume of an equivalent of 80 μg of this sample was adjusted to 360 μL with the thiourea/urea solution. Next, 40 μL of a 10-fold concentrated reswelling solution was added containing 2 M thiourea, 8 M urea, 10% Nonidet P-40, 200 mM DTT, and 5% Pharmalyte (pH 3–10). This sample containing reswelling solution was used for the rehydration of IPG strips in the pH range of 3–10. 2D PAGE and gel staining were performed as described above.
Protein Identification
For the identification of proteins by mass spectrometry, protein samples of 1 mg were separated by preparative 2D PAGE, and the gels were stained with Coomassie blue R250. In-gel tryptic digestion of the excised protein spots was performed by use of a peptide-collecting device (Otto et al. 1996). A 0.5 μL peptide solution was mixed with an equal volume of a saturated α-cyano-4-hydroxy cinnamic acid solution in 50% acetonitrile and 0.1% trifluoroacetic acid (v/v), and applied to a sample template of a MALDI-TOF mass spectrometer (Voyager DE-STR, PerSeptive Biosystems). Peptide mass fingerprints were analyzed using the MS-Fit software as provided by Baker and Clausner (UCSF Mass Spectrometry Facility, San Francisco) throughhttp://prospector.ucsf.edu. For N-terminal sequencing, preparative 2D gels were blotted onto a pPVDF membrane and the membrane was stained with Coomassie blue R250. The excised protein spots were applied to an Applied Biosystems Procise Sequencer and N-terminal sequencing was performed by ChromaTec GmbH Greifswald. Identified spots in different gels were matched manually.
Western Blotting and Immunodetection
To detect OpuAC, PrsA, and QoxA, B. subtilis cells were separated from the growth medium by centrifugation (2 min, 14,000 rpm, room temperature). Proteins in the growth medium were concentrated 20-fold upon precipitation with TCA, and samples for SDS-PAGE were prepared as described previously (van Dijl et al. 1991). After separation by SDS-PAGE, proteins were transferred to a PVDF membrane (Millipore Corporation) as described by Kyhse-Andersen (1984). OpuAC, PrsA, and QoxA were visualized with specific antibodies and alkaline phosphatase-conjugated goat anti-rabbit antibodies (SIGMA) as described previously (Lemma et al. 1995; Kappes et al. 1999; Tjalsma et al. 1999).
Acknowledgments
We thank Karin Binder and Renate Gloger for expert technical assistance, the members of the Groningen and European BacillusSecretion Groups (http://www.ncl.ac.uk/ebsg) for stimulating discussions, and Dr. V.P. Kontinen, Prof. Dr. E. Bremer, and Dr. A. Kröger for providing specific antisera against PrsA, OpuAC, and QoxA, respectively. We also thank Christian Scharf for the establishment and support of mass spectrometry in the laboratory of Michael Hecker. H.A., B.V., S.O., and M.H. were supported by grants from the Deutsche Forschungsgemeinschaft (DFG), the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (BMFT), and the Fonds der Chemischen Industrie to M.H. H.T. was supported by Genencor International (Leiden, the Netherlands). J.M.v.D., S.B., and M.H. were supported in part by European Union Grants QLK3-CT-1999-00413 and QLK3-CT-1999-00917.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵4 Corresponding author.
-
E-MAIL J.M.van.Dijl{at}farm.rug.nl; FAX 31-50-3633000.
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.182801.
-
- Received February 1, 2001.
- Accepted June 14, 2001.
- Cold Spring Harbor Laboratory Press