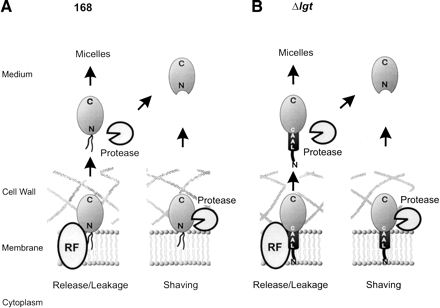
Model for the shedding of lipoproteins. (A) Shedding of the predicted lipoproteins MntA, OppA, PstS, YcdH, YclQ, YdhF, YfmC, and YqiX into the medium of the parental strain is envisaged in at least two ways. First, the lipid-modified mature lipoprotein could be passively (leakage), or actively released from the membrane. Active release would require a dedicated system (the hypothetical release factor RF), as described for lipoproteins of E. coli that are sorted to the outer membrane, or secreted into the growth medium (Yakushi et al. 2000). Thus, although there is no experimental evidence for this presently, lipid-modified mature lipoproteins could be secreted by B. subtilis in the form of micelles. Alternatively, the lipid modification of a released lipoprotein could be lost due to amino-terminal proteolysis. Second, lipid-modified mature lipoproteins could be shaved from the membrane by amino-terminal proteolysis. (B) Lipoprotein shedding by B. subtilisΔlgt can be envisaged to take place in at least two ways. First, unmodified translocated pre-lipoproteins, such as OpuAC and PrsA, could either leak from the membrane, or be actively released into the growth medium by a (hypothetical) release factor (RF). Next, the released pre-lipoproteins could form micel-like structures, or be subject to amino-terminal proteolysis. The latter would result in the presence of mature-like forms in the growth medium as observed for MntA, OppA, YclQ, YfiY, YfmC, and YxeB. Second, these unmodified pre-lipoproteins could be shaved from the membrane by amino-terminal proteolysis.











