Fine-Resolution Physical Mapping of Genomic Diversity in Candida albicans
Abstract
It has been suggested that Candida albicans, a diploid asexual fungus, achieves genetic diversity by genomic rearrangement. This important human pathogen may provide a system in which to analyze alternate routes to genomic diversity. C. albicans has a highly variable karyotype; its chromosomes contain a middle repeated DNA sequence called the Major Repeat Sequence (MRS), composed of subrepeats HOK, RPS, and RB2. RPS is tandemly repeated while the other subrepeats occur once in each MRS. Chromosome 7, the smallest of the eight chromosomes, has been previously mapped. The complete physical map of this chromosome was used to analyze chromosome 7 diversity in six strains, including two well-characterized laboratory strains (1006 and WO-1) and four clinical ones. We found four types of events to explain the genomic diversity: 1) Chromosome length polymorphism (CLP) results from expansion and contraction of the RPS; 2) reciprocal translocation occurs at the MRS loci; 3) chromosomal deletion; and (4) trisomy of individual chromosomes. These four phenomena play an important role in generating genomic diversity in C. albicans.
Candida albicans is the most common fungus responsible for oral and vaginal infection and causes systemic disease in immunocompromised hosts, such as patients who have undergone organ transplants or those undergoing intensive chemotherapy (Musial et al. 1988). In this diploid organism, for which a sexual cycle has not yet been found, evidence supports clonal propagation, at least primarily (Pujol et al. 1993; Lott et al. 1999). Despite the asexual lifestyle, C. albicans shows frequent karyotypic variation (Iwaguchi et al. 1990; Rustchenko-Bulgac 1991; Magee et al. 1992). This could be problematic in a fungus that regularly undergoes meiosis (Kistler and Miao 1992). However, in a population that undergoes clonal propagation, the diversification of karyotype will be acceptable if the progeny are viable (reviewed in Zolan 1995). Karyotypic variation has been reported in species that undergo clonal propagation (Mills and McCluskey 1990; Zolan 1995; Fierro and Martin 1999). However, the molecular mechanisms for karyotypic variation have not yet been clarified.
There are numerous reports of karyotypic diversity in C. albicans with chromosome size ranging from 4 Mbp to 0.5 Mbp (Snell and Wilkins 1986; Lott et al. 1987; Magee and Magee 1987; Lasker et al. 1989; Iwaguchi et al. 1990; Asakura et al. 1991; Rustchenko-Bulgac. 1991; Wickes et al. 1991). Suzuki et al. (1989) and Rustchenko-Bulgac et al. (1990) proposed that there was a correlation between the diversity of karyotype and diversity of phenotype. Several reports comparing karyotypic change and phenotypes–such as the ability to switch (Ramsey et al. 1994), assimilation of carbon source (Rustchenko et al. 1994 ), and resistance to the antifungal drug fluconazole (Mori et al. 1998)–have been published. Despite great effort, no correlation between the diversity of karyotype and diversity of phenotype was shown until Rustchenko and coworkers found an association between the loss or partial deletion of one of the homologs of chromosome 5 and assimilation of sorbose (Janbon et al. 1998). Subsequently, a correlation between the loss of one of the homologs of chromosome 4 and resistance to fluconazole was shown (Perepnikhatka et al. 1999).
These findings have led to the notion that gain of a new niche by karyotypic alteration leading to changes in the phenotype is an adaptation strategy of C. albicans, and the nature of the genome of this fungus has thus become an important subject. Many efforts have been made to understand the mechanism by which diversity is generated in the karyotype in C. albicans. Iwaguchi et al. (1990) and Thrash-Bingham and Gorman (1992) suggested that reciprocal chromosome translocation would account for many of the observed changes in the karyotype.
Iwaguchi et al. (1992a) cloned and characterized a repeated sequence, RPS (2 kbp), which is found on all chromosomes except chromosome 3. The number of RPS units present is dependent on both the strain and the chromosomal location. Chu et al. (1993) completed SfiI macro restriction maps of 1006 and WO-1 and showed that, in the latter strain, three reciprocal chromosomal translocations occurred near theSfiI sites. Later, it was found that the SfiI sites were highly conserved in the RPS sequences of different chromosomes (Chibana et al. 1994) and that the RPS is tandemly repeated between theSfiI fragments of chromosome 5 (Chindamporn et al. 1995) and of chromosome 7 (Chibana et al. 1998). Flanking the RPS sequences are additional repetitive elements RB2 (6 kbp) and HOK (8 kbp) (Chindamporn et al. 1998). The complete repeat unit is referred to as the Major Repeat Sequence (MRS) and is composed of HOK, RPS, and RB2, in that order (Chibana et al. 1998).
In this study, we confirm that the MRS plays a major role in the changes that lead to karyotypic diversity in C. albicans. The variations in karyotype of several clinical isolates are shown to result from translocations and chromosome length polymorphism (CLP), both involving the MRS. Thus, the RPS element serves as both a breakpoint for chromosomal translocations and, through expansion of its number of internal repeats, a major source of size variation between homologs. We have mapped karyotypic rearrangements to high resolution in several clinical isolates of C. albicans and compared them to strain 1006, the source of the mapping part of the C. albicans Genome Project (http://alces.med.umn.edu/candida.html). This study shows that the MRS is involved in CLP and supports the model that it serves as a site of ectopic pairing for recombination leading to translocations between nonhomologous chromosomes. This shows a mechanism of karyotypic rearrangement that may serve as a model for chromosomal rearrangements in a variety of fungi. We show a role of MRS for karyotypic rearrangement that may rovide one explanation for the presence of intermediate repeats in the genomes of diploid organisms and also serve as a model for chromosomal rearrangements in a variety of fungi and other eukaryotes.
RESULTS
Sizes of the Chromosomes and SfiI Fragments
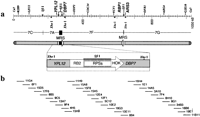
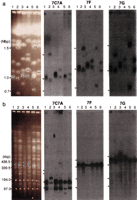
An accurate and detailed physical map of chromosome 7 in C. albicans strain 1006 has been constructed (Chibana et al. 1998). This map is based on a genomic library constructed in the fosmid vector; the average size of the C. albicans genomic DNA insert in this vector is ∼35 kbp (Kim et al. 1992). Chromosome 7 (1 Mbp in size) is the smallest of the eight homologous chromosomes and consists of four SfiI fragments, 7C, 7A, 7F, and 7G, and two copies of the MRS; the repeats form the borders between SfiI fragments 7A–7F and 7F–7G and are oriented in an inverted repeat configuration (Fig. 1a). A fosmid contig composed of the minimal number of fosmids covering the entire chromosome 7 is shown in Figure 1b. To examine the distribution of all the elements of chromosome 7 in the strains that showed CLP of chromosome 7, the minimal tiling set was divided into three smaller contigs representing the parts of the chromosome separated by MRSs (fragments 7C–7A, 7F, and 7G); these clones were pooled and the pools used as probes for the following experiments: Intact chromosomal DNAs of the strains were separated with CHEF and blotted to membranes. The membranes were hybridized with the three contig probes separately. Several positive bands were found in the strains, ranging in size from 760 kbp to 1,500 kbp (Fig. 2a). This result suggests that the elements of chromosome 7 of 1006 are dispersed among different-sized chromosomes in the other strains. The size of the chromosomes was measured and is shown in Table1. For convenience the chromosomes containing material found on chromosome 7 in 1006 will temporarily be called chromosome (a), chromosome (b) and chromosome (c) (from largest to smallest in each strain). Names reflecting their origins will be considered in the discussion. The genomic DNAs of the strains were digested with SfiI, run on CHEF gels, and blotted to membranes. The three contig probes were hybridized to the membranes (Fig. 2b). The size of the SfiI fragments that hybridized with the contig probes was measured (Table 1). Except for a few bands, the sizes of these fragments are generally conserved across the strains.SfiI fragment 7D in WO-1 is the result of an SfiI RFLP that includes fragment 7C and 7A (Chu et al. 1993). One of theSfiI fragments 7G in NUM55 was 40 kbp larger than 7G in 1006. One of the 7A fragments of NUM1000, which is only 40 kbp, will be discussed below. These were the only significant polymorphisms found among the SfiI fragments, in clear contrast to the CLP. In addition to the fosmid contigs, DNA markers (LEU2 [7C], CDC34 [7F], ARG4 [7G]; see Fig. 1a) were used as probes for the chromosomes and the SfiI fragments. There was no inconsistency between the results using the individual genes and the fosmid contigs except in NUM1000 (data not shown). The hybridization profiles were most complicated in NUM1000. Although chromosome (c) hybridized to all three fosmid contig probes, its size was only 760 kbp. The hybridization signal with the 7C–7A contig was much weaker than with chromosome (b) in NUM1000 (Fig. 2a), and one of the SfiI fragments, 7A, was much smaller than in other strains (Fig. 2b and Table 1). This suggests that a chromosomal deletion has occurred. To confirm this hypothesis, chromosome (c) in NUM1000 was eluted from the gel, labeled with 32P, and hybridized to blots of the DNAs of the minimal fosmid contig. The result of this experiment indicates that all of the 7C and half of the 7A fragments were deleted from chromosome (c).
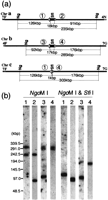
Physical maps of DNA markers on chromosome 7 in Candida albicans strain 1006. DNA markers and restriction maps: The original maps were reported by Chibana et al. (1998). DNA markers used in this study are shown on the top. Fragments 7A, 7F, and 7G are separated by MRSs that include SfiI sites. Fragments 7C and 7A are separated by an SfiI site that is not in an MRS. The two MRSs are inverted and flanking SfiI fragment 7F. OnlyXhoI sites adjacent to MRS are shown. The MRS region between 7A and 7F was magnified to show the order of subrepeat elements RB2, RPS and HOK (a). Minimal fosmid contig: The minimal contig is composed of the minimum number of fosmids covering the entire chromosome 7. The minimal contig was divided into three contigs, 7C–7A, 7F, and 7G, separated by the two MRS sequences, to identify the distribution of chromosome 7 elements among the different strains. The fosmids from 6F1 to 13A7 were used as the 7C–7A contig, those from 15H9 to 13E11 as the 7F contig, and those from 16H4 to 34B3 as the 7G contig. Fosmids 11G4, 11B11, 19E1, and 18B6 include subtelomeric repeats 9F4, 4H5, and 8B4 include the MRS (b).
Hybridization of the fosmid contiguous sequence (contig) probes to intact chromosomes and to SfiI fragments. The Southern hybridization profiles with fosmid contig probes 7C–7A, 7F, and 7G are shown. The gels on the left are marked with colored dots that show which bands correspond to the hybridization profiles on the right. Yellow, red, and blue dots indicate gel bands that hybridize with the 7C–7A, 7F, and 7G contigs, respectively. Karyotypes of six different strains are shown: lane 1, 1006; lane 2, WO-1; lane3, NUM55; lane 4, NUM114; lane 5, NUM1000; lane 6, 1719. Hybridization to intact chromosomes: The three bars at the left of each image indicate 1.5 Mbp, 1.0 Mbp, and 0.7 Mbp, respectively. The size was estimated with intact chromosomes ofSaccharomyces cerevisiae and Lambda ladder DNAs (a). Hybridization to SfiI restriction fragments: The four bars at the left of each image indicate 436.5 kbp, 339.5 kbp,194.0 kbp, and 97.0 kbp, the sizes of Lambda ladder DNAs (b).
Size of Chromosome 7 and SfiI Fragments
MRS Size
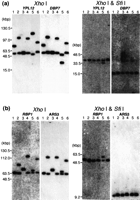
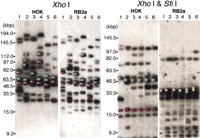
The element within the MRS that is tandemly repeated, RPS, contains several SfiI sites separated by 0.2 kbp to 1.1 kbp (Chibana et al. 1994). Because there may be several copies of RPS and because there are two MRSs on chromosome 7, it seemed possible that variations in the MRS might contribute significantly to the CLP of chromosome 7. To measure the size of the gaps between the SfiI fragments 7A-7F and 7F-7G, the genomic DNA was digested with the enzyme XhoI, which does not cut in the MRS of chromosome 7. The fragments were then separated by pulse-field gel electrophoresis, blotted, and probed with markers that map close to the boundaries of the two MRSs in 1006. The size of the fragment that hybridized with the probes gives a measure of the size of the intact MRS. As shown in Figure 1a, YPL12,DBP7, RBP1, and ARS3 are close to the MRSs and hybridized to SfiI fragments 7A, 7F, 7F, and 7G respectively, in all strains. Figure 3a shows the results with the sequences YPL12 and DBP7. The two probes hybridized to the same band in all strains except NUM1000. The size of each XhoI fragment was measured (Table2). The genomic DNA was also subjected to double digestion with XhoI and SfiI, separated by contour-clamped homogeneous electric field (CHEF), and blotted to a membrane that was once again probed with YPL12 andDBP7 (Fig. 3a). To measure the size of gap between theSfiI fragments 7A and 7F, the sizes ofXhoI-SfiI fragments were subtracted from the size ofXhoI fragment (Table 2). The size of the gap between theSfiI fragments 7F and 7G was calculated in the same way, using the RBP1 and ARS3 probes and is shown in Figure 3b and Table3. These two probes hybridized to same band in all strains except WO-1.
Southern hybridization with probes flanking the MRS to restriction fragments of XhoI and XhoI–SfiI digests. Because the MRSs in chromosome 7 do not include XhoI sites, aXhoI digest leaves fragments including the entire MRS sequences and the adjacent DNA. Four MRS flanking markers,YPL12, DBP7, RBP1, and ARS3 were included in the XhoI fragments (Fig. 1). The genomic DNA of each strain was digested with XhoI and with XhoI andSfiI. The DNA was separated by countour-clamped homogeneous electric field and blotted onto a membrane. Lane 1, 1006; lane2, WO-1; lane 3, NUM55; lane 4, NUM114; lane5, NUM1000; lane 6, 1719. The DNA size was determined with the MidRange 1 PFG marker (New England Biolabs) and 1 kb ladder (Bethesda Research Laboratories). The MRS between 7A and 7F: probesYPL12 and DBP7 (a). The MRS between 7F and 7G: probesRBP1 and ARS3 (b).
Measurement of the Gap between SfiI Fragments 7A and 7F
Measurement of the Gap between SfiI Fragments 7F and 7G
Location and Orientation of the MRSs
All the XhoI-digested bands that had been identified withYPL12 and DBP7 were also positive with RB2 and HOK (Fig. 4). These bands hybridized with RPS as well (data not shown). All of the XhoI-SfiI double-digested bands that hybridized to YPL12 were positive with RB2 but not with HOK, while all of the XhoI-SfiI double-digested bands that were positive with DBP7 were also positive with HOK but not with RB2 (Fig. 4). RPS did not hybridize with any of the bands in the double digests (data not shown). These results show that the DNA markers YPL12, RB2, RPS, HOK, andDBP7 occur in that order at the 7A–7F junction on both homologs in 1006, WO-1, NUM55, NUM114, and 1719 and on one homolog in NUM1000. For the 7F and 7G junction, the same experiment was carried out (Fig. 4), showing that the DNA markers RBP1, HOK, RPS, RB2, and ARS3 exist in that order at the 7F–7G junction on both homologs in 1006, NUM55, NUM114, NUM1000, and 1719 and on one homolog of WO-1.
Orientation of the MRS with respect to the flanking markers. The membranes shown in Figure 3 were stripped and rehybridized with RB2a (a part of RB2) and HOK. The bands shown to hybridize to the flanking markers are indicated by colored circles. The bands hybridized to the MRS flanking markers YPL12 (yellow), DBP7 (red),RBP1 (green), and ARS3 (blue) in Figure 3 indicated by dots on the hybridization profiles.
Translocations in WO-1 and NUM1000
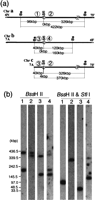
A reciprocal translocation was found between chromosomes 7 and 4 (Chu et al. 1993) in WO-1. To determine the origins of chromosomes (a) and (b) in NUM1000, the bands corresponding to these chromosomes were isolated from a pulse-field gel, labeled, and probed to a blot of the fosmid library. Chromosome (a) hybridized to the fosmids previously assigned to fragments 7F and 7G but not to those from 7C and 7A and also to fosmids from chromosome 4 (data not shown). Chromosome (b) in NUM1000 hybridized to the fosmids of 7C–7A but not those of 7F and 7G and also to fosmids from chromosome 4 (data not shown). This result indicates that chromosomes (a) and (b) contain translocation products of chromosomes 7 and 4. To investigate whether the translocations in NUM1000 and WO-1 occurred between the MRSs of chromosomes 7 and 4, we looked for MRS-flanking markers on chromosome 4. Using sequence data of the Candida genome project (http://www-sequence.stanford.edu/group/candida/), DNA markers flanking other MRSs of unknown location in the genome were prepared by PCR. To find DNA markers flanking the MRS of chromosome 4, whole DNA of chromosome 4 from strain 1006 was labeled and hybridized to the PCR products. We identified four markers from chromosome 4: OGH3 and OGH12, adjacent to HOK, and OGR4 and OGR9, adjacent to RB2 (data not shown). OGH3 hybridized to SfiI fragment 4F, and OGR9 hybridized to fragment 4H. The sizes of the SfiI fragments of chromosome 4 in WO-1 and NUM1000 are shown in Table 4. To investigate the size of the MRS and the connecting RPS fragments,XhoI-SfiI mapping as in Figure 3 was carried out with OGH3, OGH12, OGR4, and OGR9. In both strains, OGH3 and OGR9 were assigned to the same XhoI band, confirming the connection between 4F and 4H, and the size of RPS cluster was determined (Table5). There are extra XhoI sites between the markers and the MRS on SfiI fragment 4N, soNgoMI- SfiI mapping rather than XhoI-SfiII mapping was performed to examine the junction of the translocation between chromosomes 4 and 7 in WO-1. RBP1, which is adjacent to HOK on 7F, and OGR4, which is adjacent to RB2 on 4N, were all assigned to a 235 kbp band (Fig.5) before digestion with SfiI and were found on separate fragments after. We concluded that the order of markers on this fragment of chromosome (a) is RBP1, HOK, RPS, RB2, and OGR4. Similarly, OGH12, which is adjacent to HOK on 4F, and ARS3, which is adjacent to RB2 on 7G, were assigned to the same 285 kbp band (Fig. 5) and were separated after digestion withSfiI I; thus, OGH12, HOK, RPS, RB2, and ARS3, in that order, define the translocation junction of chromosome (b).
Size of SfiI Fragments of Chromosome 4
Measure of the Gap between SfiI Fragments 4F and 4H
Junction of the translocation between chromosomes 4 and 7 in WO-1.NgoMI and SfiI restriction map of the junction (a). The locations of the MRS flanking markers are indicated by numbers in circles: (1) RBP1; (2) OGR4; (3) OGH12; and (4) ARS3. The locations of RBP1 and ARS3 in chromosome 7 are shown in Figure 1. OGR4 and OGH12 were assigned to SfiI fragments 4N and 4F of chromosome 4, respectively (not shown). Southern hybridization with markers flanking the MRSs of chromosomes 4 and 7 (b). Genomic DNA of WO-1 was digested with NgoMI and withNgoMI and SfiI and resolved as described in Methods. The fragments were then transferred onto membranes and hybridized withRBP1, lane 1; OGR4, lane 2; OGH12, lane3; and ARS3, lane 4. OGH12 shows weak cross-hybridizaton to 7F. The DNA size was determined by Lambda ladder DNA markers.
In the translocation between chromosomes 4 and 7 in NUM1000 there is an extra XhoI site and an NgoMI site between the markers, so BssHII-SfiI mapping was performed to examine the junction of the translocation. OGR4, which is adjacent to RB2 on 4E, and DBP7, which is adjacent to HOK on 7F, were assigned to the same 422 kbp-band (Fig. 6) and were separated by SfiI digestion. Thus, markers at the junction of chromosome (a) in NUM1000 are, in order, OGR4, RB2, RPS, HOK, and DBP7. YPL12, which is adjacent to RB2 on 7A, and OGH12, which is adjacent to HOK on 4F, were similarly assigned to a 160 kbp band (Fig. 6) and separated with SfiI. YPL12, RB2, RPS, HOK, and OGH12 are located at the junction of chromosome (b) in NUM1000, in that order.
Junction of the translocation between chromosomes 4 and 7 in NUM1000.BssHII and SfiI restriction maps of the junction (a). The locations of the MRS flanking markers are indicated by numbers in circles: (1) OGR4; (2) DBP7; (3) YPL12; and (4) OGH12. The locations of YPL12 and DPB7 in chromosome 7 are shown in Figure 1. OGR4 and OGH12 were assigned to SfiI fragments 4N and 4F of chromosome 4, respectively (not shown). Southern hybridization with MRS flanking markers of chromosomes 4 and 7 (b). Genomic DNA of WO-1 was digested with BssHII and withBssHII and SfiI. The restriction fragments were separated by contour-clamped homogeneous electric field. The fragments were then transferred onto membranes and hybridized with OGR4, lane1; DBP7, lane 2; YPL12, lane3; and OGH12, lane 4. The DNA size was determined by Lambda ladder DNA markers.
Structures of Translocation Products
We used the information derived from the previous sections to deduce the structure of the translocation chromosomes in WO-1 and NUM1000. Chromosome (a) in WO-1 includes SfiI fragments, 7C, 7A, 7F, 4N, and 4BB. Because 4BB, which is adjacent to 4N, hybridized to the telomere sequence, it must be located at one of the ends of the chromosome, arbitrarily written here as the right-hand end. The OGR4 probe is located at the left end of 4N and hybridizes with a 235 kbpNgoMI fragment (Fig. 5). The RBP1 probe is at the right end of 7F and hybridizes with the 235 kbp NgoMI fragment. The gap between 4N and 7F is 18 kbp. The probe DBP7, which is at the left end of 7F, and the probe YPL12, which is at the right end of 7A, hybridized with a 105 kbp XhoI fragment. The gap between 7F and 7A is 49 kbp (Table 3). Adjacent to 7C, which includes telomere sequences at the left end, is 7A. Therefore, this chromosome is composed of fragments 7C (170 kbp)-7A (90 kbp)-RPS (49kbp)-7F (340 kbp)-RPS (18 kbp)-4N (830 kbp)-4BB (33 kbp) (Table 6). The total calculated size is 1,530 kbp, close to the measured size of chromosome (a), 1,540 kbp. The same analysis was applied to other chromosomes and other strains. A summary of the structures of the chromosomes containing part or all of chromosome 7 is shown in Table 6.
Total Size of the SfiI Fragments and Gaps
Aneuploidy of Chromosome (b) in NUM1000
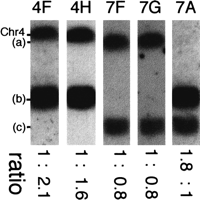
Although chromosome (b) has SfiI fragments in common with chromosomes 4, and (c) in NUM1000 (Fig.7)7, the probes assigned to chromosome (b) always hybridized with a higher intensity to this band than to other chromosomes. For this reason, we investigated the ploidy of chromosome (b) in NUM1000. The strength of the signal with the probes OGH12 and OGH9 on chromosome (b) is about twice that of chromosome 4, which is monosomic (Fig. 7). Probe YPL12hybridizes to chromosome (b) with an intensity approximately twofold that of chromosome (c), which is monosomic. The signals on chromosomes (a) and (c) with DBP7 and ARS3 are very similar. These results indicate that chromosome (b) is disomic, so 7C and part of 7A are disomic, and the telomere-proximal part of 7A and all of 4F and 4H are trisomic in NUM1000.
Aneuploidy of chromosome (b) in NUM1000. The chromosomal DNAs of NUM1000 were separated and transferred onto a membrane. The membrane was hybridized to OGH12, OGR9, DBP7, ARS3 and YPL12, and markers were assigned to 4F, 4H, 7F, 7G, and 7A respectively. The numbers at the bottom show the intensity ratios.
Correspondence between the Calculated Size of the MRS and Intensity of Hybridization of RPS
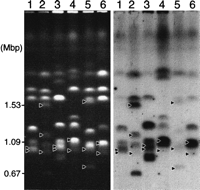
The sizes of the RPS clusters in the two MRSs in chromosome (a) in 1719 and the NUM114 were 74 kbp and 101 kbp. On the other hand, the total size of the RPS cluster is only 1 kbp on chromosome (a) in NUM1000 (Table 6). To confirm that the size of the RPS cluster calculated from the restriction mapping results reflects the number of RPSs, we hybridized Southern blots of pulse-field chromosome separations with an RPS probe (Fig.8). The intensities of hybridization of the chromosome 7 homologs and chromosome 7-related chromosomes (Fig. 8, indicated by arrowheads) were correlated with the calculated size of the RPS cluster. In NUM114 and 1719 the intensity of chromosome (a) was much higher than the intensity of chromosome (b), indicating that two large MRSs exist on chromosome (a).
Hybridization of chromosomes with the RPS probe. Lane 1, 1006; lane 2, WO-1; lane 3, NUM55; lane 4, NUM114; lane 5, NUM1000; lane 6, 1719. Arrowheads indicate chromosomes (a), (b), and (c) in each lane. Lane 1, 1006; lane2, WO-1; lane 3, NUM55; lane 4, NUM114; lane5, NUM1000; lane 6, 1719.
DISCUSSION
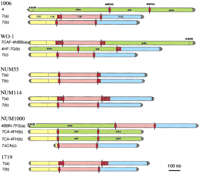
Much effort has been expended to understand the nature of the genome and its apparent plasticity in C. albicans. In this study, we analyzed the physical basis of the size variation of chromosome 7 in six strains that show significant polymorphism of this chromosome. We show here that the great majority of changes involve the MRS; this result suggests that this intermediate repeat may play a leading role in the production of karyotypic variability in C. albicans, including both size variation of intact homologs and translocations involving nonhomologous chromosome. The mechanism of translocation we propose involves crossing over between nonhomologous chromosomes due to pairing at the homologous region of itself on different chromosomes. Such a model makes some testable predictions. The strongest prediction is that the directionality of the MRS dictates allowed translocation products (Fig. 9). That is, translocation can only occur between fragments on opposite ends of the repeat. Thus, we would not expect a fragment on the RB2 side of the MRS to end up in a translocation with another fragment at that side. Figure 9 shows that the translocations we have studied have been the result of permitted recombination events. The second prediction is that translocation should be rare or nonexistent at SfiI sites that do not involve the MRS. This prediction is weaker because it can only be supported by absence of data. However, in the cases studied here, both these predictions are fulfilled.
Schematic representation of the chromosomes. Telomeric and subtelomeric regions are shown by vertical stripes on the ends. The colored boxes indicate 7C7A (yellow), 7F (pink), and 7G (blue). Green boxes indicates translocated SfiI fragments of chromosome 4. Red arrows represent the sizes and orientation of the MRSs, in the order (head to tail) of HOK, RPS, and RB2.
MRS Mediates Translocations
The MRS, which includes the sequence elements HOK, RPS, and RB2, in that order (Chibana et al. 1998; Chindamporn et al. 1998), is located on seven of the eight chromosomes; only RB2 is found on chromosome 3 (Chindamporn et al. 1998). RPS includes several SfiI sites (Iwaguchi et al. 1992a; Chibana et al. 1994). The two MRSs on chromosome 7 in 1006 are inverted and flank the 350 kbp SfiI fragment, 7F (Chibana et al. 1998). Chu et al. (1993) reported a reciprocal translocation of SfiI fragments between chromosomes 7 and 4 in WO-1. In this study, a similar but not identical translocation was found in NUM1000. The reciprocal translocation in WO-1 occurred by recombination between the MRS at 4F–4N and that at 7F–7G. The translocation in NUM1000 occurred at the MRSs at 4F–4N and 7A–7F. Iwaguchi et al. (2000) found a strain that has an extra chromosome composed of SfiI fragments, 4H, 7F, and 7G. We found that the MRS occurs between 4F and 4H on chromosome 4 and that a reciprocal translocation between the MRS at 7A–7F and the MRS at 4F–4H most likely occurred. Navarro-Garcia et al. (1995) reported a reciprocal translocation of SfiI fragment 7G and 2A. A translocation between MRS of 7F–7G and of 2U–2A is proposed to have led to that chromosome. Chromosome 2 is composed of SfiI fragments 2U and 2A (Chu et al. 1993). This suggests that an MRS exists between these two elements, which was found (H. Chibana, in prep.). It seems likely that any MRS region is a potential site for translocations.
Conservation of Synteny in Fragments Bounded by the MRS
Although some polymorphisms independent of the MRS were found in the strains studied here, the size of the SfiI fragments was mostly conserved. Iwaguchi et al. (1992a) reported that among fifty clinical isolates, the size of chromosome 3, which lacks HOK and RPS, was the most conserved. Our data, together with that observation, suggest that translocation is mediated by the MRS in the most cases. Thus, we propose that C. albicans translocation chromosomes be named to reflect SfiI synteny. Chromosomes in which no translocation is found, such as chromosome 7 in 1006, NUM55, NUM114, and 1719, would be called chromosome 7 as before. To distinguish homologous chromosomes of differing sizes, the larger one would be chromosome 7a and the smaller 7b. In WO-1, we call chromosome (a) chromosome 7CAF-4NBB, chromosome (b) is chromosome 4HF-7G, and chromosome (c) is chromosome 7. In NUM1000, chromosome (a) is designated Chromosome 4BBN-7FG, chromosome (b) is chromosome 7CA-4FH, and chromosome (c) is chromosome 7DCA (Fig. 9).
MRS Mediates Chromosome-Length Polymorphism Not Involving Translocations
CLP due to change in the number of rDNA repeats has been reported inC. albicans (Iwaguchi et al. 1992b; Rustchenko et al. 1993). Variation in the number of repeats is a common feature of tandemly repeated DNA, as we have shown with the RPS element here. The MRS includes a subunit, RPS, that is tandemly repeated (Iwaguchi et al. 1992a). We have observed that changes in the number of copies of RPS can change the size of a chromosome homolog in several of the strains. The sizes of the RPS clusters in the two MRSs in one of the homologs of chromosome 7 in 1006, 1719, and NUM114 were 26 kbp, 74 kbp, and 101 kbp and the sizes of the chromosomes were 1030 kbp, 1080 kbp, and 1120 kbp, respectively (Table 6). In NUM114, about 10% of chromosome 7a is composed of the MRSs, including several tandem repeats of RPS.
Chromosome-Size Polymorphisms Not Involving the MRS
Chu et al. (1993) reported that C. albicans strains WO-1 and 1006, which have different karyotypes, show highly conservedSfiI restriction maps. In this study, the sizes of theSfiI fragments of chromosome 7 were mostly conserved in the six strains studied, although they possess widely different sizes of chromosome 7. An exception occurred in NUM1000, in which chromosome 7c has lost all of fragment 7C and two-thirds of fragment 7A. Size polymorphisms of less than 10 kbp are found more often in SfiI fragments 7C and 7G, which are telomeric, than in the 7A and 7F fragments (Table 1). In NUM55, one of the 7G fragments is 40 kbp larger than 7G in 1006 (Table 1). There was no evidence of translocation in this SfII fragment. McEachern and Hicks (1993) reported telomere-length polymorphism in C. albicans. Louis (1994)reported polymorphism in the subtelomeric regions in Saccharomyces cerevisiae. Care2 (Lasker et al. 1992) and Rel-2 (Thrash-Bingham and Gorman 1993), which are repeated genomic elements, were assigned to the subtelomeric regions on chromosome 7 fragments C and G of strain 1006 (Chibana et al. 1998). Thus, one possible reason for the size variation of these fragments in other strains is length polymorphism in the telomere or in subtelomeric repeats. An alternative mechanism for length polymorphism of the SfiI fragments is the insertion or excision of transposons. Goodwin and Poulter (2000) have examined the emerging sequence of the C. albicans genome and have found evidence for more than 350 transposon insertions. Variations in gene size have been attributed to the excision of retrotransposons 1006 and CAI-4 (J.L. Beckerman, H. Chibana, J. Turner, and P.T. Magee, in prep.), and their presence or absence would affect chromosome size in unrelated strains.
Natural Aneuploidy in Candida albicans
Aneuploid strains of C. albicans have been reported since the early 1980s (Whelan and Magee 1981). If a strain has lost one homolog of a chromosome, it grows slowly (Barton and Gull 1992; Magee and Magee 1997; Janbon et al. 1998). Janbon et al. (1998) found that selection for growth on L-sorbose resulted in loss of one homolog of chromosome 5. Growth on glucose selects for strains that have two copies of the remaining homolog. The same group later showed that the loss of one homolog of chromosome 4 is associated with resistance to fluconazole in one Candida strain (Perepnikhatka et al. 1999). It is possible that there are other conditions that select strains that have undergone chromosome nondisjunction. One of these unknown conditions may have selected NUM1000, which has undergone nondisjunction of a translocation chromosome, chromosome 7A–4F.
Molecular Basis of Genome Fingerprinting with Ca3, 27A, and RPS
Ca3 (Sadhu et al. 1991), 27A (Scherer and Stevens 1988), and RPS (Iwaguchi et al. 1992a) probes have been used as tools for epidemiological analysis of C. albicans infections. When genomic Southern blots of various strains are probed with any of these sequences, restriction fragment-length polymorphisms are found (Pujol et al. 1997). Ca3, 27A, and RPS are included in the MRS, and all share some common sequences (Chindamporn et al. 1998; Chibana et al. 1998). What is the basis of the variation revealed by these probes? A major cause is the diversity in the size of RPS unit due to the number of short periodic internal repeats in found in different strains (Chibana et al. 1994; Doi et al. 1998). A second reason is heterogeneity in sites for the restriction enzyme EcoRI. Sites for this enzyme, which is used in many epidemiological studies, are not highly conserved in the RPS family (Chibana et al. 1994), so that EcoRI cuts the whole RPS cluster out occasionally. As shown in Figures 8 and 9, the number of RPSs varies among MRSs, chromosomes, and strains. The combination of all these factors leads to different profiles of hybridization with Ca3, 27A, and RPS. Sister chromatid exchange and interchromosomal gene conversion presumably operate to change the number of RPS repeats and the specific sequence of some of the RPSs at MRSs (Pujol et al. 1999). These changes result in the diversity in the Southern-hybridization profiles of strains as they diverge from a common ancestor. The results of this study show that the genome ofC. albicans is unique among the fungi so far analyzed. Its linkage unit is not a full chromosome, as is the case with the great majority of eukaryotes; instead the genome is constructed of gene clusters bound by a complex intermediate repeat, the MRS. Conservation among strains is at the level of these clusters, which rearrange at a relatively high frequency to yield karyotypes that, although usually diploid in gene content, do not contain pairs of identical homologs.
The organism that seems to lack a sexual cycle but is able to adapt to a variety of niches, as shown by its normally commensal but sometimes parasitic lifestyle, C. albicans must have mechanisms to provide genetic variation. Chromosome rearrangement has been proposed as one such mechanism (Rustchenko-Bulgac et al. 1990; Chu et al. 1993), and this study provides both the evidence for this rearrangement and a proposed mechanism. Lacking is the demonstration that the rearrangements we describe here affect gene expression. The imminent completion of the physical map (Chibana et al. 1998) and of the sequence (http://www-sequence.stanford.edu/group/candida/) will provide the information necessary to investigate differences in gene regulation related to chromosome rearrangement.
METHODS
Strains
The 1006 strain (Goshorn and Scherer 1989) has been used as standard for chromosome analysis in C. albicans. This strain was chosen because it possesses eight pairs of homologous chromosomes, each pair of homologs almost identical in size except for chromosome R (Chu et al. 1993). The electrophoretic karyotype of 1006 is the one most frequently found in clinical isolates. WO-1 (Slutsky et al. 1985) was shown by Chu et al. (1993) to have several reciprocal translocations, including one involving chromosome 7. Iwaguchi et al. (1990) selected ten strains showing chromosome length polymorphism (CLP) from 50 clinical strains in Nagoya, Japan. Of the ten strains, three—NUM55, NUM114, and NUM1000, all of which showed CLP in chromosome 7—were chosen for this study. The 1719 was chosen from five clinical isolates in the Scherer strain collection (http://alces.med.umn.edu/Candida/strainlist) because it also showed CLP in chromosome 7.
Polymerase Chain Reaction (PCR)
Oligo DNAs, shown below, were synthesized and used for PCR amplification of the DNA markers:
HIS4, 5′CCTCAGTCACAATAAGGACTGGACGAT3′ and 5′CGGGGTGAGATTGGCTGATATTGAGAA3′; OGH3, 5′CCCACTTAATTTCCCACATCCAGTTACTT3′ and 5′CGG TAACCATTTGTTAAAAACAATTGTATC3′; OGH12, 5′AATC TAGCCCGATGTGGAAAACCAT3′ and 5′CACATCACCAACT TAGCACCTTCATT3′; OGR4, 5′TTGTAAACTGTGAATTG CAGTTGATGAGTT3′ and 5′GTCATAGTAGTAATATGA TGTGTGGCGTGC3′; OGR9, 5′AGAAGGAGGTACACAATG GACCATT3′ and 5′CTAAGGAACCACGAGGGTGAAAGTA3′; RBP1, 5′GCATCGTGGGGATACTAACTAACTAC3′ and 5′TG CAGCTATATATCGAGGTTTTAGTTCAAC3′. PCR products were amplified with each primer pair. All PCR was carried out under the following conditions: Step 1, 94°C for 3 min; step 2, 94°C for 30 sec; step 3, 55°C for 30 sec; step 4, 70°C for 1 min; step 5, repeat step 2, thirty times; step 6, 68°C for 10 min with 2.5 M MgCl2.
DNA Probes and Labeling
All probes used in this study are shown in Figure 1. Preparation of fosmid DNA and whole-chromosome DNA was described by Chibana et al. (1998). PCR products and restriction fragments were purified by agarose gel electrophoresis (Heery et al. 1990). The isolated DNA was labeled with 32P-dCTP using the RediPrime kit (Amersham).
Southern Blots and Hybridization
The chromosomal DNAs separated by pulse-field electrophoresis were transferred onto Hybond-N+ membrane (Amersham). Hybridization to the probes was carried out with 50% (v/v) formamide, 6x SSPE, 5x Denhardt's solution and 0.5% of SDS at 42°C for overnight. The membrane was washed twice with 2x SSPE and 0.5% SDS at room temperature for 15 min and once with 0.1x SSPE and 0.5% SDS at 65°C for 30 min.
Pulse-Field Separations
DNA preparation for pulse-field gels essentially was as described by Chu et al. (1993). Separation of chromosomes was carried out at 15°C in 0.5X TBE (44.5 mM Tris, 44.5 mM Boric acid, 1 mM EDTA) on a BioRad CHEF (contour-clamped homologous electric field) DRIII instrument. Chromosome size between 1.1 Mbp and 1.6 Mbp was estimated using chromosomal DNA of S. cerevisiae (Bio Rad). Lambda ladder PFG Markers (New England Biolabs) were used as size markers in the 50-kbp–1,100-kbp range, Mid Range DNA size markers (NEB) for 15 kbp to 400 kbp, and a 1-kb ladder (BRL) in the 12-kbp-and-below range. The conditions for separation are as follows: (Fig. 2a) 0.9% agarose gel, 80 sec to 130 sec, 5 V/cm, 120° included angle for 24 h, 150 sec to 300 sec, 4.5V/cm, 120° for 16 h, and then 720 sec to 900 sec, 2.0V/cm, 106° for 24 h. (Fig. 2b) 0.9% agarose, 7 sec to 100 sec, 4.5 V/cm, 120° for 24 h, and then 80 sec to 400 sec, 3.5V/cm, 120° for 24 h. (Fig. 3a) 0.9% agarose gel, 6.0 V/cm, 120° for 14 h, and then 8 sec to 30 sec, 5 V/cm, 120° for 6 h. (Fig. 3b) 0.9% agarose gel, 0.5 sec to 4 sec, 6.0 V/cm, 120° for 14 h, and followed by 3 sec to 20 sec, 4 V/cm, 120° for 5 h. (Fig. 5) 0.9% agarose, 5 sec to 30 sec, 6 V/cm, 120° for 14 h, and followed by 30 sec to 80 sec, 5 V/cm, 120° for 8 h. (Fig. 6) 0.9% agarose, 7 sec to 100 sec, 4.5 V/cm, 120° for 24 h, and then 80 sec to 400 sec, 3.5 V/cm, 120°, for 12 h. (Fig. 8) 0.7% agarose III (Amresco), 70 sec to 100 sec, 5 V/cm, 120° for 12 hr, 100 sec to 300 sec, 4.5 V/cm, 120° for 26 h, and then 720 sec to 900 sec, 2.0 V/cm, 106° for 16 h.
Densitometry
The densitometry of each bands of Figure 7 was performed with the program NIH Image. The ratio of bands on each lane was calculated after subtraction of the background which was measured on each lane.
Acknowledgments
We thank Ikuyo Mizuguchi for sending strains NUM55, NUM114, NUM1000, and the rest of the NUM series and Suzanne Grindle for strain 1719. This work was supported by Grants No. AI16567 and AI35109 from the National Institute of Allergy and Infectious Diseases.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵1 Corresponding author.
-
E-MAIL chibana{at}biosci.cbs.umn.edu; FAX (612) 625-5754.
-
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.148600.
-
- Received May 23, 2000.
- Accepted September 22, 2000.
- Cold Spring Harbor Laboratory Press