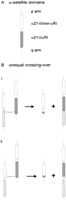Extreme Reduction of Chromosome-Specific α-Satellite Array Is Unusually Common in Human Chromosome 21
Abstract
Human centromeres contain large arrays of α-satellite DNA that are thought to provide centromere function. The arrays show size and sequence variation, but the extent to which extremely low levels of this DNA can occur on normal centromeres is unclear. Using a set of chromosome-specific α-satellite probes for each of the human chromosomes, we performed interphase fluorescence in situ hybridization (FISH) in a population-screening study. Our results demonstrate that extreme reduction of chromosome-specific α satellite is unusually common in chromosome 21 (screened with the αRI probe), with a prevalence of 3.70%, compared to ≤0.12% for each of chromosomes 13 and 17, and 0% for the other chromosomes. No analphoid centromere was identified in >17,000 morphologically normal chromosomes studied. All of the low-alphoid centromeres are fully functional as indicated by their mitotic stability and binding to centromere proteins CENP-B, CENP-C, and CENP-E. Sensitive metaphase FISH analysis of the low-alphoid chromosome 21 centromeres established the presence of residual αRI as well as other non-αRI α-satellite DNA suggesting that centromere function may be provided by (1) the residual αRI DNA, (2) other non-αRI α-satellite sequences, (3) a combination of 1 and 2, or (4) an activated neocentromere DNA. The low-alphoid centromeres, in particular those of chromosome 21, should provide unique opportunities for the study of the evolution and the minimal DNA requirement of the human centromere.
The centromere is a specialized structure on a chromosome that plays an essential role in chromosomal segregation during mitosis and meiosis. The most abundant DNA element in the human centromere is the α satellite, which constitutes as much as 3%–4% of chromosomal DNA. The significance of α satellite in centromere function has been the subject of active research for over two decades. Its apparent ubiquitous presence at the cytogenetically defined primary constrictions of all normal human chromosomes implies a priori a possible functional role. Analysis of rearranged (Tyler-Smith et al. 1993) or fragmented (Brown et al. 1994) centromeres of the human Y chromosome has demonstrated that sequences necessary for centromere function are localized to a region containing 150–200 kb of α-satellite DNA. Five different studies involving the introduction of exogenous α satellite into cultured mammalian cells have provided further evidence in favor of α-satellite DNA having a functional centromere role. In three of these studies, transfection of α satellite into hamster, simian, or human cells has led to the appearance of integration-associated centromere manifestation such as increase in the number of dicentric and ring chromosomes per cell (Heartlein et al. 1988) or the formation of a primary constriction, disruption of normal chromosome segregation, and association with the α-satellite DNA-binding protein CENP-B (Haaf et al. 1992; Larin et al. 1994). In the remaining two studies, transfection of α-satellite DNA with other genomic DNA, in particular human telomere sequences, into human cell cultures has resulted in the formation of stable minichromosomes containing functional centromeres that can be attributed to the introduced α-satellite DNA (Harrington et al. 1997; Ikeno et al. 1998).
Although the above evidence points strongly to a positive centromere role for α satellite, it is now clear that this DNA is not always, on its own, sufficient to elicit centromere activity. For example, in human dicentric chromosomes, normal amounts of α satellite are present on two distinct chromosomal sites and yet only one of these sites will form an active centromere (Earnshaw et al. 1989; Page et al. 1995; Sullivan and Schwartz 1995). Furthermore, an increasing number of functional neocentromeres have been detected in marker chromosomes that are formed from interstitial chromosomal sites that are devoid of α-satellite sequences (for review, see Choo 1997b), suggesting that α-satellite DNA is, in certain situations, unnecessary for centromere activity. A well-characterized example of such neocentromeres is the one derived from band q25.2 of human chromosome 10 in which the core centromeric protein binding region has been localized to a genomic sequence of ∼80 kb (Voullaire et al. 1993; du Sart et al. 1997; Cancilla et al. 1998; Barry et al. 1999).
Structurally, centromeric α-satellite DNA is composed of huge arrays of tandemly repeated monomers of 171 bp (Waye and Willard 1987;Choo et al. 1991). Arrays of α satellite on different chromosomes are highly heterogeneous both in the sequence of the monomer as well as in their higher-order organization. Such heterogeneity has led to the evolution of chromosome-specific subfamilies of α satellite within the human genome, in which a particular subfamily may be specific to a single chromosome or shared by two or more chromosomes, or where multiple subfamilies of this DNA may be present on a single chromosome (for review, see Choo et al. 1991; Choo 1997a).
In addition to sequence heterogeneity, α-satellite arrays also demonstrate extensive size variation between nonhomologous as well as homologous chromosomes. This variation has been recognized at both the molecular and cytogenetic levels. Using pulsed-field gel electrophoresis (PFGE), significant differences in the size of individual α-satellite DNA arrays have been observed between homologs of various human chromosomes, such as the X chromosome (array size range: 1380–3730 kb; Mahtani and Willard 1990), Y chromosome (array size range: 285–1020 kb; Tyler-Smith 1987; Abruzzo et al. 1996), or chromosome 21 (array size range, 420 to >2650 kb; Marcais et al. 1991; Trowel et al. 1993; Ikeno et al. 1994). Other studies of individual chromosomes 1 (Waye et al. 1987b), 3 (Waye and Willard 1989), 7 (Wevrick et al. 1992), 8 (Ge et al.1992), 10 (Jackson et al. 1993), 11 (Waye et al. 1987a), 13 (Trowell et al. 1993), 14 (Trowell et al. 1993), and 16 (Greig et al. 1989) have indicated a broad centromeric α-satellite size spectrum ranging from 200 kb to ∼4 Mb. At the cytogenetic level, interphase and metaphase FISH analyses have similarly revealed noticeable variations in α-satellite array signals in functionally normal centromeres (e.g., Shelby et al. 1996;Verma et al. 1997). Although these studies have unequivocally established the quantitative heteromorphic nature of α-satellite DNA, it is unclear to what extent extreme reductions of these arrays are tolerated in the normal centromeric regions of different human chromosomes. In the present study, we addressed this question in a population study employing FISH and a battery of chromosome-specific α-satellite DNA probes for each of the human chromosomes. Our results indicated that extreme reduction of this DNA is rare for the different chromosomes with the exception of chromosome 21. A similar analysis was performed on a cohort of Down syndrome (DS) patients. The implications of our observations in the light of the evolution and minimal DNA requirement of α satellite, and the etiology of meiotic nondisjunction in DS, are discussed.
RESULTS
The study was divided into two phases. The first involved the use of each of the α-satellite probes to determine the distribution profile of cells that exhibited the expected number of FISH signals within a sample population. In the second phase, a more rapid interphase FISH screening of a larger population sample was undertaken with the specific aim of identifying extreme cases of α-satellite DNA reduction on each of the chromosomes.
Phase I: Determination of FISH Profiles for Different α-Satellite Probes
Table 1 lists the different chromosome-specific α-satellite probes used in the present study. Most of the probes are specific for a single chromosome and are expected to give two predominant interphase FISH signals corresponding to the homologous chromosomes. However, because of the evolution of common α-satellite sequences on a number of the nonhomologous chromosomes, three of the probes hybridize to the centromeres of more than one homologous chromosome pair, giving four expected FISH signals for αRI (chromosomes 13 and 21) and αXT (chromosomes 14 and 22) probes, and six expected signals for pZ5.1 (chromosomes 1, 5, and 19). In initial experiments, we assessed the suitability of each probe to detect its cognate chromosomes on a small number of cell lines by comparing the signals obtained in interphase nuclei with those observed in metaphase preparations. This analysis demonstrated the existence of a good correlation between interphase and metaphase FISH signals for all of the probes (except pZ5.1) and established the validity of using these probes in our interphase FISH screening procedure. The pZ5.1 was found to be relatively unreliable because of the uneven intensity of signals for the different nonhomologs (with those for chromosome 19 being particularly weak) as well as the increased difficulty of scoring six interphase nuclear signals. As such, the pZ5.1 probe was applied only to metaphase preparations in the second phase of the study.
Interphase FISH Screening for Low-Alphoid Cell Lines
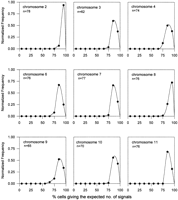
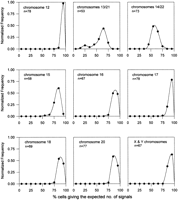
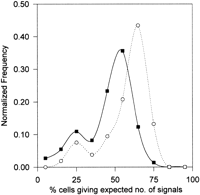
Cell lines (50–80) from our fixed-cell repository that have been studied previously by routine cytogenetics and shown to be normal were used for the determination of interphase FISH profiles. This involved scoring an average of 100 interphase nuclei per cell line per probe to determine the percentage of cells giving the expected number of FISH signals relative to the total number of cells scored. For each probe, using this percentage as a characteristic parameter for each cell line, all of the cell lines studied are grouped and categorized into a relative frequency distribution curve as shown in Figure1. As can be seen, for all of the monochromosomal-specific probes, a single peak was observed. This represents the predominance of a high percentage of cells (>70%) within each of these cell lines giving the expected number of signals. A single distribution peak was similarly seen with the αXT probe (chromosome 14/22) except that the distribution peak is somewhat broader and shifted to the left. Such differences can be attributed to the fact that simultaneous hybridization of four chromosomes has increased spatial crowding, leading to an underestimation of the expected interphase signals. Control experiments were performed on a similar panel of cell lines using a combinations of probes specific for chromosomes 12 and 17 (pB12 and TR17, respectively) to recreate artificially the situation of scoring four signals (Fig. 2, solid squares). These chromosomes, when investigated separately, previously have demonstrated a single distribution peak of a high percentage of cells giving the expected two signals (Fig. 1). However, when in combination, they also revealed a left shift in a broader distribution profile. A substantial proportion of the cells in the same cell lines were shifted towards giving lower number of signals than expected. Similar results were also reproduced using a different combination of probes, p4n1/4 + pMR9A, specific for chromosomes 4 and 9, respectively (data not shown). This demonstrates clearly that this phenomenon is not related to the properties of individual probes but is an inherent problem of the physical constraints of counting signals within a confined space.
Relative frequency distribution curves of interphase FISH performed with chromosome-specific α-satellite probes (see Table 1) on cytogenetically normal subjects. Horizontal axes represent the percentage of cells in each cell line showing the expected number of interphase FISH signals. Data points represent the mid-values of the bins (in the range of 10%) used for the construction of the frequency distribution diagram. Vertical axes denote the number of cell lines in each bin expressed as a fraction of the total number (n) of cell lines investigated.
Relative frequency distribution curves of interphase FISH performed to recreated artifically the scoring of four (●) and five (◼) signals on cytogenetically normal control subjects. Data points represent the mid-values of the bins (in the range of 10%) used for the construction of the frequency distribution diagram. The horizontal axis represents the percentage of cells within a cell line giving the expected number of signals. The vertical axis denotes the number of cell lines in each bin expressed as a fraction of the total number of cell lines investigated for each of the reconstruction experiments. Four signals were produced using a combination of probes, pB12 and TR17, specific for chromosomes 12 and 17, respectively (n = 112). Five signals were produced using a combination of the same probes together with the Y chromosome probe, pLAY5.5, on male subjects (n = 86). The data for chromosome 17 (▴) from Fig. 1 as an example of scoring two signals were reproduced for comparison.
A somewhat different result was obtained with the αRI probe (chromosome 13/21), in which two overlapping subpopulations of cell lines were discernible. As shown in Figure 1 (chromosomes 13/21), the majority of the cell lines gave the expected number of signals for this probe, with a distribution peak that was closely similar to those obtained for the αXT probe and the artificial four-signals situations. However, a smaller but significant peak was detected in the lower range, representing cases in which only a disproportionately low percentage (<35%) of cells gave the expected number of signals. This minor peak, which constituted 6 of the 53 cases, was unlikely to be the result of technical artifact associated with scoring four interphase FISH signals as it was not observed in the frequency distribution profile for αXT and the control experiments. A more likely explanation is that the cell lines in the minor peak carried one or more chromosomes that have lower-than-normal amount of α satellite on their centromeres, rendering a reduced probability of detection at interphase FISH under the experimental conditions specified. That this was the case was established by direct FISH analysis of metaphase chromosomes, which has indicated the presence of a chromosome in each of these cell lines with a greatly reduced (but not zero; see below) amount of αRI DNA. Such an analysis has further revealed that the low-alphoid chromosome in every case corresponded to chromosome 21 and not chromosome 13 despite the fact that the chromosomes were equally recognized by the αRI probe. These results formed the basis for the phase II study involving a significantly larger sample size.
Phase II: Population Screening for Low-Alphoid or Analphoid Centromeres
For this screening, we used an additional >300 human lymphoblastoid cell lines that have been established as part of our ongoing tissue culture collection for patients and related individuals that have presented with a variety of clinical interests. Formal cytogenetic examination was performed on these cell lines and those known to carry chromosomal abnormalities were excluded from the present study. High-stringency interphase FISH was performed on these cell lines using each of the chromosome-specific α-satellite DNA probes (except pZ5.1). For pZ5.1, metaphase preparations were examined visually for centromeres that gave FISH signals that were significantly smaller than those generally observed on chromosomes 1, 5, or 19. With the other probes, ∼50 interphase nuclei were screened and cell lines were characterized by a predominance of cells exhibiting signals significantly less than the expected number based on the distribution profiles depicted in Figure 1 (i.e., <35% for αRI and αXT, and <70% for the remaining probes) were identified for further investigation by metaphase FISH analysis.
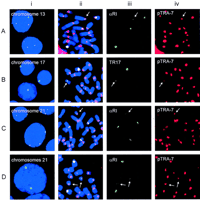
The outcome of the screening is summarized in Table 1. Extreme low-alphoid/analphoid centromeres were noted in only three of the human chromosomes. A single cell line was identified for each of chromosome 13 (Fig. 3A, i) and chromosome 17 (Fig. 3B, i) which, when the data of phases I and II studies were combined, gave a chromosomal prevalence of 0.12% and 0.11%, respectively. In stark contrast, for chromosome 21, 24 cell lines showed extremely low αRI in one of the homologs (an example is shown in Fig. 3C, i), whereas in one cell line both of the homologs were affected (Fig. 3D, i). When these results were combined with those obtained in the phase I study, a chromosomal prevalence of 3.70% was obtained for chromosome 21 (Table1). Using simple χ2 tests in 2 × 2 tables for a comparison of the result for chromosome 21 to those for other chromosomes, a P value < 0.001 was shown in each case. It can, therefore, be concluded that low-alphoid centromere is unusually common in chromosome 21.
Interphase and metaphase FISH analysis of low-alphoid centromeres for chromosome 13 (A), chromosome 17 (B), and chromosome 21 (C,D). The cell line in C contains a single low-αRI chromosome 21; the cell line in D contains two low-αRI chromosomes 21. (i) Typical interphase FISH appearances; (ii) dual-color FISH using a combination of high-stringency chromosome-specific α-satellite probing (green) and low-stringency nonspecific α-satellite probing (red); (iii) split images of ii showing fluorescein isothiocynate (FITC) signals for the chromosome-specific α-satellite sequences; (iv) split images of iishowing Texas Red signals for the nonspecific α-satellite sequences. Nuclei and chromosomes were counterstained with DAPI and pseudocolored blue. α-Satellite probes used to detect chromosomes 13/21, 17, and nonspecific α-satellite DNA were αRI, TR17, and pTRA-7, respectively. Arrows indicate the low-alphoid centromeres.
No Analphoid Chromosomes Were Detected
We were further interested to know (1) whether the low-alphoid chromosomes were analphoid, and (2) whether other α-satellite sequences not recognized by the specific probes used were present in the low-alphoid centromeres. To address the first question, metaphase FISH was used in conjunction with enhanced image analysis using a charge-coupled device (CCD) video camera. The results indicated the presence of a detectable TR17 signal on the chromosome 17 case (Fig.3B, ii,iii), and low but discernible αRI signals on all of the affected chromosomes 13 and 21 (Fig. 3A, ii,i; C, ii,iii; D, ii,iii). These results indicated that analphoid centromeres do not exist in our sample series. Further support for this came from results obtained in addressing the second question.
For the determination of whether other α-satellite sequences not recognized by the specific alphoid probe used were present in the low-alphoid centromeres, high-stringency FISH was first performed using the chromosome-specific α-satellite probes on metaphase chromosomes to clearly identify the affected homolog. This was followed by low-stringency FISH using the pTRA-7 probe to detect all α-satellite sequences. Figure 3B (ii–iv) show that for chromosome 17, relatively little difference was observed between the high-stringency and low-stringency hybridization signals on the low-alphoid centromere, suggesting that only a small amount of non-TR17-related α-satellite sequence was present on this centromere. In contrast, all of the low-alphoid centromeres for chromosome 13 (Fig. 3A, ii–iv) and chromosomes 21 (Fig. 3, C and D, ii–iv) showed a noticeably stronger signal at low stringency compared to high stringency. These results indicated that the low-alphoid centromeres of chromosomes 13 and 21 contained a substantial amount of non-αRI-related α-satellite DNA.
Low-Alphoid Centromeres Bind Key Centromere-Specific Antigens
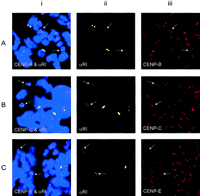
The functional status of the low-alphoid centromeres was determined by immunocytochemical studies using antibodies specific for the centromere proteins CENP-B, CENP-C, and CENP-E (see Discussion). Figure4 shows results obtained for the cell line shown in Figure 3D where both of the chromosome 21 homologs demonstrated markedly reduced amounts of α-satellite DNA. As can be seen, specific signals corresponding to all three antigens were detected on both of the low-alphoid chromosomes 21 in this cell line. The colocalization of these antigens with the FISH signals for the αRI probe suggested the centromere activity is located within or very close to the αRI DNA domain. When the immunocytochemical/FISH analysis was performed on all of the other low-alphoid cases for chromosomes 13, 17, and 21 identified in this study, similar results to those presented in Figure 4 were obtained.
Combined immunocytochemical and FISH analyses for the detection of CENP-B (A), CENP-C (B), and CENP-E (C) with the αRI probe. The cell line contains two low-αRI chromosomes 21 (arrows). (i) Combined images of centromeric protein signals (red) and the αRI signals (green); (ii) split images of i showing the FITC signals for the αRI domain; (iii) split images of i showing the Texas Red signals for the centromeric proteins. Nuclei and chromosomes are counterstained with DAPI and pseudocolored blue.
Quantitation of the Low-αRI Arrays
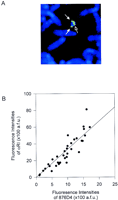
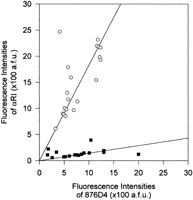
The sizes of the chromosome 21-specific αRI arrays were estimated using Quantitative-FISH (Q-FISH). The amount of fluorescence attributable to each of these arrays was compared directly to the signals for the 876D4 YAC clone containing unique genomic sequences of a known size (570 kb) located on the same chromosome. We first validated our method in a somatic cell hybrid WAVR-4d-F9-4a with a human chromosome 21. The αRI signals and 876D4 signals can easily be distinguished from each other by their respective cytogenetic locations (Fig. 5A). A plot of the fluorescence intensities of αRI signals against 876D4 signals of the somatic cell hybrid is shown in Figure 5B. A similar plot of the fluorescence intensities for the αRI signals against the 876D4 signals for a typical cell line containing one low-αRI chromosome 21 is shown in Figure 6. It is evident from these plots that the absolute values of the α satellite and 876D4 signals were variable among homologs within individual cells and between cells of a particular cell line. This may be because of differences in chromatin condensation among individual cells or even between homologous chromosomes within a single cell. Alternatively, differences in the microenvironments on a slide may result in variable probe accessibility or hybridization outcome. This is also clearly shown in Figure 5B where the fluorescence signals of a unique genomic sequence on the single chromosome 21 in stable culture can be variable among metaphase spreads on the same slide. Based on these considerations, it will not be prudent to compare fluorescence signals measured in different cells or on different homologs. In contrast, a relatively uniform microenvironment will be expected for signals that coexist on a particular homolog. That this is so is evident from the linear relationship that exists between the α satellite and the intrachromosomal 876D4 signals (Figs. 5B and 6). Furthermore, the distribution profiles shown in Figures 5B and 6 indicate a greater scattering of data for the normal centromeres (open circles) than those for the low-alphoid centromeres (solid squares). This is probably due to overextension of the signals for the normal centromeres beyond the dynamic range of the digital image capture algorithm. In addition, the average size of αRI array in a typical centromere is known to be in the megabase order (Marcais et al. 1991; Trowell et al. 1993; Ikeno et al. 1994), which greatly exceeds the size of the 876D4 genomic segment (570 kb) to which αRI is compared. The gross differences in signal intensities of the two signals render an optimal capture and an accurate comparison impractical. The low-alphoid centromere, in contrast, appears to be well within both the dynamic and size ranges. Based on the premises that ∼40% interpersed repeat sequences are present on any given unique genomic sequence (Smit 1996), that most of these repetitive DNA have been competed away during the prehybridization step with Cot-I DNA (see Methods), and that a linear relationship exists between the αRI and 876D4 signals, the size of the αRI array, s, can be estimated using the formula s = a × 0.6 × 570 kb, wherea is the regression coefficient as determined by linear regression of data for the low-alphoid centromere of each individual cell line. Using this calculation, the sizes of the αRI arrays of the somatic cell hybrid WAVR-4d-F9-4a were estimated to be 1.33 Mb and those for the low-alphoid chromosome 21 identified in the Phase II study ranged from 51 to 184 kb with a mean of 78 kb.
Q-FISH analysis of the centromeric αRI domain and the 876D4 genomic region at 21q22.1–22.2 in the human chromosome 21 somatic cell hybrid WAVR-4d-F9-4a. (A) The 876D4 signals (solid arrows) on the long arms of sister chromatids of chromosome 21 can easily be distinguished from the centromeric α-satellite signals (open arrow). (B) Each point represents the fluorescence intensities of αRI and 876D4 probe on the human chromosome 21 in different metaphase spreads on the same slide. Data are fitted to a straight line by linear regression. (a.f.u.) Arbitrary fluorescence units.
Q-FISH analysis of the centromeric αRI domain and the 876D4 genomic region at 21q22.1–22.2 in a cell line containing one low-alphoid chromosome 21. Each point represents the fluorescence intensities of αRI and 876D4 probe on a particular chromosome; (○) a normal-alphoid chromosome 21; (◼) a low-αRI chromosome 21. Each data set shows a good fit to a straight line by linear regression. (a.f.u.) Arbitrary fluorescence units.
Prevalence of Low-Alphoid or Analphoid Chromosome 21 in DS
The prevalence of low-alphoid or analphoid centromeres in patients with DS was investigated. Seventy-three cases derived from the same fixed-cell repository and matched for the period of −20°C storage with the control samples used in Phase I study were analyzed by interphase FISH. All of the DS cases have previously been cytogenetically shown to consist of three free chromosomes 21. In the present study, a systematic interphase FISH analysis of these cases using each of the chromosome-specific α-satellite probes listed in Table 1 did not detect any new case of low-alphoid or analphoid centromeres except for the αRI probe.
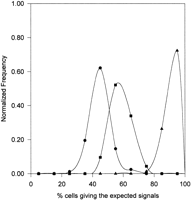
The results of interphase FISH performed with the αRI probe under high stringency were scored as described above for phase I study. The frequency distribution curves in Figure 7 indicated that, as with the control data (Fig. 1, chromosomes 13/21), two subpopulations of cell lines were observed, with the bulk of the cell lines giving the expected five signals in a distribution peak that was closely similar to, albeit slightly shifted to the left of, that seen in the controls. As with the control subjects, a smaller second peak was seen in the DS population. Reconstruction experiments aimed at producing five signals in the male subjects of the control and DS cell lines using four combinations of probes—(1) TR17 + pB12 + pLAY5.5, (2) TR17 + pB12 + pLAX, (3) αXT + pLAY5.5, and (4) αXT + pLAX—revealed, in each case, a single distribution peak similar to that seen with the αXT probe alone (Fig. 1, chromosomes 14/22) and the artificial four signals (Fig. 2, solid squares) but further shifted left (Fig. 2, solid circles) supporting the theory of spatial signal crowding. These distribution profiles suggested that the appearance of the smaller second peak in the DS samples was not artifactual and was likely to be related to the presence of a significant proportion of cell lines in which the amount of αRI on chromosomes 13 and/or 21 was drastically reduced. Confirmation of this came from metaphase FISH, which indicated that 15 of the low-αRI cases involved a chromosome 21, whereas in two cases a chromosome 13 was involved. These results gave a prevalence of 6.85% (based on three chromosomes 21 per DS case) for chromosome 21, and 1.37% for chromosome 13. Employing a simple χ2 test, the number of low-αRI centromeres in chromosome 21 in DS samples showed a noticeable difference from that of the normal population at the 5% significance level (P = 0.0419). A similar level of significance was obtained for chromosome 13 between the DS and control samples (P = 0.0121). As in the control study, metaphase FISH has indicated that all of the affected centromeres have retained a small but detectable amount of αRI. Low-stringency hybridization further revealed that these centromeres also contained a substantial amount of non-αRI alphoid sequences. The fixed samples were not suitable for centromeric protein detection and this was therefore not done.
Relative frequency distribution curves of interphase FISH using the probe αRI performed on a cohort of 73 DS cell lines (◼). (○) The results derived from Fig. 1 of the control group using the same probe. Horizontal axis represents the percentage of cells in each cell line showing the expected five interphase FISH signals for DS cells. Data points represent the mid-values of the bins (in the range of 10%) used for the construction of the frequency distribution diagram. Vertical axis denotes the number of cell lines in each bin expressed as a fraction of the total number of cell lines investigated.
DISCUSSION
In this study we have performed a population screening using FISH and a battery of chromosome-specific α-satellite probes for all of the human chromosomes to determine the prevalence of centromeres showing extreme reductions of the α-satellite DNA. Because the study focuses specifically on the centromeric DNA found on the normal primary constriction, it has not included dicentric chromosomes or analphoid marker chromosomes exhibiting neocentromeric activities. As shown in Table 1, between 300 and 450 cases (representing 600–900 diploid chromosomes, except for the X and Y chromosomes) were analyzed. The results indicated that extreme reduction of this DNA, with a low probability of detection at interphase, is a rare occurrence within the normal human genome, with a prevalence of 0.12% and 0.11% for each of chromosomes 13 and 17, and 0% for the remaining chromosomes. A notable exception is chromosome 21 where extreme reduction of the chromosome-specific αRI α-satellite array is relatively common (3.70%) in comparison with the other human chromosomes. Using metaphase FISH coupled with a highly sensitive detection system based on a cooled CCD camera with digital image enhancement, we have further ascertained that residual α-satellite sequences are present on all of the affected chromosomes. Thus, in total, the analysis of ∼17,000 different human chromosomes has not revealed any analphoid centromere. This outcome contrasts those of earlier studies that have reported a number of analphoid chromosomes 21 based on a relatively small number of cases (Verma and Luke 1992; Weier and Gray 1992; Verma et al. 1997). We believe the discrepancy is likely to be due to the lower sensitivity of the other investigator's methods, which were generally based on interphase and metaphase FISH using conventional fluorescence microscope detection. Furthermore, unlike our study, which has compared all of the human chromosomes, previous investigators have analyzed only one or two chromosomes (Wevrick et al. 1990; Mizunoe and Young 1992;Verma and Luke 1992; Weier and Gray 1992; Bossuyt et al. 1995; Abruzzo et al. 1996; Verma et al. 1997; Liehr et al. 1998; Verma et al. 1998) and therefore do not provide a relative measure of low-alphoid centromeres among the different chromosomes.
The centromere of human chromosome 21 (which has a similar organization to that of human chromosome 13; Trowell et al. 1993) is divided into two broad domains: the α21-I domain, which consists of a relatively homogeneous array of αRI sequences, and the α21-II domain, which contains a collection of heterogeneous, non-αRI-related α-satellite sequences (Fig. 8A; Trowell et al. 1993; Ikeno et al. 1994). Previous studies have demonstrated that the α21-I/αRI domain is rich in CENP-B-box DNA motifs and binds an abundance of CENP-B proteins, whereas the α21-II/non-αRI domain is CENP-B-box-poor and does not bind CENP-B significantly (Ikeno et al. 1994). Transfection experiments using human telomere-retrofitted YACs containing either the α21-I/αRI or α21-II/non-αRI domain have shown that the former was able to induce the formation of stable minichromosomes in a cultured human cell line, whereas the latter was not (Ikeno et al. 1998; Masumoto et al. 1998). These studies suggest that α21-I/αRI, rather than α21-II/non-αRI, serves to provide centromere function. It is, therefore, interesting that despite the greatly reduced level of α21-I/αRI in our low-αRI chromosomes 21 (and the single case of chromosome 13), the affected centromeres have remained fully functional in mitosis. That this is the case is indicated by several lines of evidence. Cytogenetic analysis has indicated complete stability of all of the affected chromosomes in the patients of origin and, for the samples used in the Phase II study, also in long-term tissue culturing. The detection of CENP-B binding on the low-alphoid centromeres suggests that sufficient CENP-B-box-containing α-satellite DNA has remained on these centromeres although CENP-B binding per se is not necessarily indicative of, nor immediately associated with, centromere activity, as this antigen is detected on both the active and inactive centromeres of human dicentric chromosomes (Earnshaw et al. 1989; Page et al. 1995;Sullivan and Schwartz 1995), and targeted disruption of Cenpbin mice is not accompanied by any apparent impairment of centromere activities (Hudson et al. 1998; Kapoor et al. 1998; Perez-Castro et al. 1998). Direct proof of centromere function in the low-alphoid chromosomes has come from immunocytochemical studies using antibodies against two functionally important centromere proteins, CENP-C and CENP-E. CENP-C has been reported to regulate kinetochore structural integrity (Tomkiel et al. 1994) and to be essential for mitosis as evident from a lethal phenotype in gene knockout studies (Fukagawa and Brown 1997; Kalitsis et al. 1998), whereas CENP-E is a molecular motor that has been shown to move chromosomes along microtubules (Thrower et al. 1995). The importance of CENP-C and CENP-E in centromere function is further suggested by the observation that these proteins are found on active, but not inactive, centromeres in dicentric or multicentric chromosomes (Earnshaw et al. 1989; Page et al. 1995; Sullivan and Schwartz 1995; Faulkner et al. 1998). Thus, the detection of both of these antigens on each of the low-alphoid centromere cases has provided direct support for the functional integrity of these centromeres.
(A) The general organization of α-satellite domains on chromosome 21 (and chromosome 13); (B) proposed model for the formation of low-alphoid centromere by unequal crossing-over caused by a misalignment of the α-satellite arrays between two homologous α21-I/αRI domains (i) and between two nonhomologous α21-I/αRI and α21-II/non-αRI domains on different chromosomes 21 (ii).
Using a novel Q-FISH procedure involving comparison of fluorescence intensities between an αRI array and an intrachromosomal unique sequence, we have estimated the lower limit of the size of the naturally occurring α21-I/αRI domain in our series. The αRI array size of the chromosome 21 in the somatic cell hybrid WAVR-4d-F9-4a estimated by Q-FISH is 1.33 Mb, which is slightly smaller than the genomic fragment detected in PFGE (1.65–1.70 Mb). This provides the validation of the technique of quantitation of FISH signals using an internal standard. This method allows simultaneous measurement of the standard and the sequence of interest, providing the internal correction of systemic variations in signal excitation and emission as well as instrumental variability. The choice of an internal standard in the vicinity of the subject of measurement further minimizes the variability of microenvironment innate to chromosome condensation state and stages of cell cycle as well as local differences on the slides. The target of our interest is the α satellite, which is a highly repeated array; the ideal internal standard should be another known repetitive sequence with monomer of similar size. This will also allow control for differences in hybridization kinetics. The use of a YAC as the internal standard suffers from the drawback that it is dependent on the richness ofAlu repeats within the YAC to be a true representation of the length of genomic sequence and an extra prehybridization step, which is not required for α-satellite probing. 876D4 in a gene-rich region of the long-arm of chromosome 21 seems to fulfill partially some of these requirements although it is not repetitive in nature. The estimated size of the αRI array with Q-FISH is of the same order of magnitude as published data. The observed discrepancy may be interpreted in light of two facts. On the one hand, the strong α-satellite FISH signals may have exceeded the dynamic range of the microscope system as evident by the greater scattering of the data as compared with that of low-αRI centromere (Figs. 5B and 6). The big contrast of intensities of the normal alphoid signal and the YAC signal also renders optimal quantitative capture of both signals impossible. On the other hand, sequence analysis has indicated divergence of the αRI sequences at the boundary region of α21-I (Mashkova et al. 1998). Under the high-stringency conditions used in our experiments, these diverged sequences may not be recognized by our probe. Furthermore, it is not feasible to attempt to detect these sequences by lowering the stringency of FISH as this would result in cross-hybridization to the α21-II domain. However, as discussed above, our method is capable of recognizing and estimating the size of the more uniform subset of αRI monomers that by far predominates the α21-I domain and has been implicated as the functional component of the chromosome 21 centromere (Ikeno et al. 1994, 1998;Masumoto et al. 1998). Because the αRI probe detects four chromosomes (two chromosomes 13 and two chromosomes 21), conventional methods of array-size estimation involving PFGE or fiber-FISH studies would yield highly complex patterns that are difficult to interpret. Only by individually transferring the chromosomes with low-alphoid centromeres to a mouse or hamster background (see, e.g., Shen et al. 1998) can these measurements and a more detailed sequence analysis be performed more confidently.
Q-FISH analysis of our low-αRI centromeres from chromosomes 21 has indicated a mean size of 78 kb, with the smallest of these measuring 51 kb. Previous reports based on PFGE analysis on a small number of chromosome 21 in monochromosomal somatic cell hybrid backgrounds have described the sizes of αRI domain ranging from 420 kb to substantially >2 Mb (Marcais et al. 1991; Trowell et al. 1993; Ikeno et al 1994). Thus, if centromeric function is provided by the αRI sequences (Ikeno et al. 1994, 1998; Masumoto et al. 1998), our data suggest that as little as 50–100 kb of this DNA may be all that is needed. It is also possible that centromeric functions in the low-αRI chromosomes may be supported by alternative mechanisms involving the non-αRI alphoid sequences or an activated neocentromere that may exist in the vicinity of the normal primary constriction site. The possibility of triggering one or more of these alternative mechanisms may explain the observed higher tolerance of chromosome 21 against a severe reduction of the αRI domain. Further studies aimed at discriminating between these possibilities should provide useful information on the minimal centromere DNA requirements as well as the possible existence of alternative functional centromeric domains on this chromosome.
The tandemly repeated nature of α-satellite sequences and the apparently nonmosaic status of the low-alphoid chromosomes in our subjects suggest a model for the formation of these centromeres through unequal meiotic crossing-over (Smith 1976). Such an unequal crossing-over presumably occurs in meiosis I and involves the mispairing of tandem arrays of α-satellite DNA between the two homologs. Depending on the site of mispairing and subsequent crossing-over, gross imbalance in the amount of α satellite may occur in the resulting products (Fig. 8B). In the case of the low-αRI centromeres of chromosomes 13 and 21, mispairing and exchange presumably occur predominantly between homologous αRI arrays (Fig. 8B, i) where maximal sequence recognition is possible and where the exchange does not result in the loss of the α21-II/non-αRI domain, as our low-stringency FISH data suggest. However, the less likely possibility that the misalignment may involve αRI array on one homolog and non-αRI α-satellite array on the other homolog cannot be excluded (Fig. 8B, ii). Recombination at the centromeric region is generally considered a rare event although this phenomenon has been studied in a limited number of chromosomes and has not been extended specifically to chromosome 21 (Mahtani and Willard 1998; Puechberty et al. 1999). Nevertheless, recombination events with unequal crossing-over has been suggested as a possible mechanism to explain the observed sequence divergence within α-satellite arrays in chromosome 17 (Waye and Willard 1986) and chromosome 21 (Mashkova et al. 1998). Our observation that low-alphoid centromeres are very rare in chromosomes other than chromosome 21 is on the one hand consistent with these published data but on the other hand suggests the possibility that recombination within the centromere of chromosome 21 may be comparatively more frequent. In addition to the unequal crossing-over model, other mechanisms such as looping out or deletion hot spots that result in the loss of centromere α-satellite DNA may also be possible.
The analysis of a cohort of 73 DS patients has revealed α-satellite distribution patterns that are closely similar to the control group for each of the probes. In particular, we wanted to determine the prevalence of low-αRI centromeres in the DS chromosome 21. The results indicated a prevalence of 6.85%, which is somewhat higher than the 3.70% for the control group. It is unclear whether the difference is sufficiently significant to suggest low-αRI status as a contributing factor in the etiology of DS although, even if so, the magnitude of the observed difference suggests that it is unlikely to be a major factor. Similarly, whether the observed increase in the prevalence of low-αRI in chromosome 13 in DS (1.37% vs. 0.12% in the controls) has any etiological significance, such as interchromosomal effect involving direct recombinational exchange between the homologous centromere domains of chromosomes 13 and 21 (Choo 1990), is unknown at present. More detailed analyses involving segregation studies and determination of the parental origin of the mis-segregated chromosome 21 will be necessary to provide further insight into any potential contribution low-alphoid centromeres may have in the etiology of DS. Nonetheless, it is interesting to speculate on the events that could occur during meiosis involving two homologous chromosomes with dramatically imbalanced amounts of the αRI DNA. Could the centromeres of these homologs be more prone to mispairing or be subjected to uneven microtubule capture and forces of separation, or could the residual αRI and the other non-αRI alphoid sequences be sufficient to ensure proper pairing and segregation? The answers to these questions should enlighten us on the centromeric requirement of the αRI and non-αRI alphoid sequences in the processes of mitotic and meiotic segregation both in normal and in DS cases.
METHODS
FISH analysis was performed on interphase nuclei using a set of chromosome-specific α-satellite probes, which together covered all of the human chromosomes (Table 1; for review, see Choo et al. 1991;Choo 1997a). FISH results were determined using a standard epifluorescence microscope and, where indicated, were extended with metaphase FISH using a more sensitive cooled CCD video camera system equipped with digital image enhancement capability.
Cell Lines
Control samples for phase I study and those from patients with DS were obtained from the Cytogenetic Laboratory of the Victorian Clinical Genetics Service (Melbourne, Australia). The control samples were tested to be cytogenetically normal. For the DS cases, only those with a diagnosis of 47, +21 were used to avoid confusion with atypical DS that resulted from Robertsonian translocation or partial translocation of 21q. The samples were prepared by standard cytogenetic techniques and stored at −20°C in methanol/acetic acid fixative (vol/vol 3:1). For Phase II study, established human lymphoblastoid cell lines were used. These were maintained by the Tissue Culture Laboratory of the Murdoch Institute (Melbourne, Australia). These cell lines were immortalized from peripheral blood and stored frozen under liquid nitrogen. Only those with no noticeable chromosomal abnormalities were used. De-identified code names (which were constructed from the original tissue culture serial numbers) of the patients from which the different cell lines were derived were used. These cell lines were collected over a period of 20–30 years, and based on the widely different serial numbers of these samples, it is unlikely that a substantial number of these samples were derived from the same family.
The somatic cell hybrid cell line WAVR-4d-F9-4a containing the human chromosome 21 was a gift from F.H. Ruddle (Slate et al. 1978) and was maintained under standard tissue culture conditions growing in Dulbecco's modified Eagle medium (DMEM) and 10% fetal calf serum.
Interphase FISH Screening
For phase I, archival fixed cells were sedimented at 1500 rpm (∼150g) for 10 min, followed by two washes of freshly prepared fixatives. The pellets were resuspended in a suitable volume of fixative to be spotted on slides. To facilitate more efficient screening, 20 specimens were dropped onto one slide according to a 3 × 7 grid on a standard 25 × 75-mm slide, instead of the traditional preparation of a single specimen per slide (Lo and Choo 1999).
For phase II, frozen lymphoblast cell lines unrelated to those used in phase I were retrieved from liquid nitrogen and thawed quickly in 37°C. The cell suspension (50 μl) was removed and the rest was refrozen in liquid nitrogen. Previous studies had established that acceptable viability of ∼40% could be obtained even after four successive freeze-thaw cycles (Lo and Choo 1999).
High-stringency interphase FISH was performed for the primary screening. Chromosome-specific α-satellite probes (Table 1) were used individually except for the X and Y chromosome-specific probes, which were combined so that two expected signals would be observed independent of the sex of the subject. FISH was performed as described (du Sart et al. 1997). Briefly, chromosome-specific α-satellite probes were labeled with digoxigenin by nick translation (Nick Translation Kit, Boehringer Mannheim, Castle Hill, Australia). The slides were dehydrated and denatured with 70% formamide at 70°C for 2 min. Hybridization at high stringency was done by incubation of the slides with 50 μl of hybridization mix containing 100 ng of denatured probe and 20 μg of salmon sperm DNA in 50% formamide, 10% dextran sulfate in 2× saline sodium citrate (SSC) at 37°C overnight inside a moist chamber. High-stringency wash was performed with three washes in 0.1× SSC each at 60°C for 10 min. One round of detection was generally sufficient to obtain good signals and was adopted strictly throughout the screening. Anti-digoxigenin–fluorescein, Fab fragments (Boehringer Mannheim) were used according to the manufacturer's protocol. Finally, the slides were mounted with 4,6-diamidno-2-phenylindole (DAPI, Boehringer Mannheim) in VectaShield (Vector Laboratories, Burlingame, CA) antifade solution.
Interphase FISH signals were scored by simple fluorescence microscopy strictly according to the following criteria: (1) Only nonoverlapping cells were scored; (2) signals that were close together were considered as distinct signals if they were clearly separated from one another by a distance greater than the radius of the larger signal, otherwise they were considered as a single bipartite signal; and (3) multiple focal planes were examined and the total number of signals per cell summed up. Interphase cells (50–100) were counted for each cell line. The number of cells in each cell line giving the expected number of signals was expressed as a percentage of the total number of cells scored in that particular cell line. This percentage was taken as a characteristic parameter for each cell line and was used to construct frequency distribution diagrams.
Metaphase FISH
In phase I, sufficient metaphases were generally present in each sample as these specimens were originally prepared for routine cytogenetic examinations. In a number of cell lines of interest, metaphase chromosomes were examined directly in the same slide used for the interphase FISH scoring.
In phase II, putative lymphoblast cell lines identified in the interphase screening were retrieved from liquid nitrogen and grown in RPMI 1640 supplemented with 20% fetal calf serum for at least three passages before FISH studies were performed. In contrast to the interphase FISH screening, only one cell line was dropped onto each slide to provide more cells and better spread of the metaphases for examination. High-stringency FISH was performed on these slides as described above. Metaphase images were analyzed using a Zeiss Axioskop fluorescence microscope (Carl Zeiss, Carnegie, Australia) mounted with appropriate filters to capture images with a cooled CCD video camera (Photometrics Image Point, AZ) linked to a MacIntosch PowerMac Desktop Computer System. The software, IP Lab Spectrum, version 2.5.5 (Signal Analytics Corporation, VA), was used to allow images to be acquired digitally and processed using the standard settings and scripts recommended by the manufacturer.
For cases requiring dual-color FISH for the simultaneous detection of specific and nonspecific α-satellite sequences, high-stringency FISH using the specific probe was first performed up to the washing steps. The slides were then dehydrated through an ethanol series at room temperature. FISH at low stringency to detect nonspecific α-satellite sequences was performed essentially in the same manner as high-stringency FISH, except that pTRA-7 α-satellite probe (Vissel and Choo 1991) labeled with biotin was used and hybridization was performed using 100 ng of the probe in 50 μl of 30% formamide, 10% dextran sulfate in 2× SSC with 20 μg of salmon sperm DNA, incubated overnight at room temperature inside a moist chamber. Low-stringency washing was performed with three washes of 1× SSC at room temperature. Avidin–Texas Red (Jackson ImmunoResearch Laboratory, Inc., PA) was used for immunodetection according to the manufacturer's protocol. Again, one round of immunodetection was sufficient to give good signals on all of the human centromeres.
Immunoctyochemical Analysis
Anti-CENP-B monoclonal antibody, 2D-7, was produced in pristane-primed mice as ascites fluid from the hybridoma cells purchased from American Type Culture Collection (Rockville, MD) (Hudson et al. 1998). Anti-Cenpc polyclonal antibody, Am-C1, was produced in a rabbit against a mouse Cenpc/GST-fusion product expressed in Escherichia coli. This antibody has been shown to cross-react with human CENP-C (du Sart et al. 1997). Antihuman CENP-E antibody, HX1, was a gift from T. Yen (Yen et al. 1991).
Immunocytochemical analyses were conducted on lymphoblast cultures as described previously (Page et al. 1995). Briefly, 70% confluent lymphoblast culture was treated with colchemid and hypotonically swelled as mentioned above. Cells (100 μl) at a concentration of 105 cells/ml was cytocentrifuged onto precleaned slides at 1000 rpm for 5 min (Cytospin 3, Shandon, Cheshire, UK). After brief drying at room temperature, the metaphase and interphase nuclei were treated by gentle flooding with potassium chromosome medium KCM [120 mm KCl, 20 mm NaCl, 10 mm Tris-HCl at pH 7.5, 0.5 mm Na2EDTA, 0.1% Triton X-100 (vol/vol)] and incubated for at least 10 min at room temperature. Primary antibodies diluted with 1× TEEN (1 mm triethanolamine HCl, 0.2 mm Na2EDTA, 25 mm NaCl), 0.1% Triton X-100, 0.1% BSA were then overlaid onto the slide under a parafilm coverslip and incubated for 30 min at 37°C in a moist chamber. Excess primary antibodies were then washed with three gentle floodings of 1× KB− (10 mm Tris-HCl at pH 7.5, 0.15 mNaCl, 0.1% BSA) at room temperature each for 5 min. Secondary antibodies conjugated with fluorescence dyes were incubated and washed in the same manner. High-stringency FISH using the chromosome-specific α-satellite probes were performed after the antibody staining and fixation in 10% formalin to identify the centromeres of interest. Digital images were acquired as described above.
Q-FISH
Quantitation of the low-αRI centromeres of chromosome 21 was performed on metaphase spreads using a mixture of the α-satellite probe αRI and the 876D4 probe that mapped to the long arm of chromsome 21 at q22.1–q22.2. 876D4 was purchased from Genome Systems Inc. (St. Louis, MO) as a YAC clone (Chumakov et al. 1992; Genome Database, http://www.gdb.org). Total yeast genomic DNA was prepared from a two-day culture of a single colony according to the supplier's protocol. Human-specific genomic probe was produced from the yeast DNA by Alu PCR amplification using the primers Alu-1[GGATTACAGG(C/T)(A/G)TGAGCCA] and Alu-2[5′-(A/G)CCA(C/T)TGCACTGCAGCCTG-3′] and labeled with nick translation (Archidiacono et al. 1994). FISH was performed as described with the inclusion of a prehybridization step with 20 times excess of human Cot-I DNA (Boehringer Mannheim) at 37°C for 1 hr after probe denaturation. The optimal amount of probe used per half-slide was determined to be 200 ng. Near-complete saturation of probe on its chromosomal templates was established by sequential high-stringency FISH first using the standard amount of YAC probe labeled with digoxigenin, followed by excess amount of the same probe (5 μg) labeled with biotin; the signals of biotin-labeled probe were found to be <1% that of the digoxigenin-labeled probe. Similarly, sequential FISH using different ligands has demonstrated near-complete (>99%) saturation of the chromosomal αRI templates using excess amount of αRI probe (1 μg).
Q-FISH was performed under high stringency using the 876D4 and αRI probes labeled with the same ligand (digoxigenin in most cases). Images of αRI signal at the centromere and the 876D4 signal on the chromosome 21 long arm were brought into focus in the same high-power field and captured simultaneously with the same excitation and emission filter sets. The two signals can be easily distinguished based on their respective cytogenetic locations. Raw data of the captured gray scale image were analyzed. A small area on the image containing the αRI and 876D4 signals on the same homolog was first selected. A “segmentation” function provided in the IP Lab software was used to display a standard pixel-value histogram of the selected area. The function allows interactive delineation of the area for each signal based on the same gray scale pixel value. “Fluorescence intensity” is defined as the sum of the values in each pixel within the delineated area and assigned as arbitrary fluorescence unit (a.f.u.).
Acknowledgments
The anti-CENP-B, anti-Cenpc, and anti-CENP-E antibodies are gifts from D.F. Hudson, P. Kalitsis, and T. Yen, respectively. The human chromosome 21 somatic cell hybrid is a gift from F.H. Ruddle. We thank the staff of the Tissue Culture Laboratory and the Cytogenetic Laboratory of the Murdoch Institute for invaluable assistance with cell culture and archival materials. A.W.I.L. receives an Overseas Postgraduate Research Award and a Melbourne Research Scholarship. M.R. is supported by Telethon grant E672. This work is supported by the National Health & Medical Research Council of Australia. K.H.A.C. is a Principal Research Fellow of the Council.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵3 Corresponding author.
-
E-MAIL choo{at}cryptic.rch.unimelb.edu.au; FAX 61-3-9348-1391.
-
- Received July 13, 1999.
- Accepted August 16, 1999.
- Cold Spring Harbor Laboratory Press