Transcription and potential functions of a novel XIST isoform in male peripheral glia
- Kevin S. O'Leary1,
- Meng-Yen Li2,
- Kevyn Jackson1,
- Lijie Shi1,
- Elena Ezhkova2,
- Bernice E. Morrow1,3 and
- Deyou Zheng1,4,5,6,7
- 1Department of Genetics, Albert Einstein College of Medicine, Bronx, New York 10461, USA;
- 2Department of Stem Cell Biology and Regenerative Medicine, Icahn School of Medicine at Mount Sinai, New York, New York 10029, USA;
- 3Departments of Obstetrics and Gynecology, and Pediatrics, Bronx, New York 10461, USA;
- 4Department of Neurology, Albert Einstein College of Medicine, Bronx, New York 10461, USA;
- 5Department of Neuroscience, Albert Einstein College of Medicine, Bronx, New York 10461, USA;
- 6Data Science Institute, Albert Einstein College of Medicine, Bronx, New York 10461, USA;
- 7Rose F. Kennedy Intellectual and Developmental Disabilities Research Center, Albert Einstein College of Medicine, Bronx, New York 10461, USA
Abstract
The XIST RNA is known for its critical roles in X Chromosome inactivation (XCI). It is thought to be expressed exclusively from one copy of the X Chromosome and silence it by recruiting various chromatin factors in female cells. In this study, we find XIST expression in male peripheral glia after integrated analyses of single-cell RNA-seq data from multiple human tissues and organs. Single-cell epigenomic data further indicate that the expression is likely driven by an alternative promoter at the end of the first exon, resulting in at least one shorter transcript (referred to as sXIST) that is active in Schwann cells and, moreover, at a higher level in nonmyelinating Schwann cells. This promoter exhibits similar activity in female glia. Multiple lines of evidence from bulk transcriptomic and epigenomic data from peripheral nerve tissues further support these findings. Genes coexpressed positively and strongly with sXIST in male glia show functional enrichment in axon assembly and cilia signaling, with many of them sharing putative miRNA binding sites with sXIST, whereas the negatively correlated genes are enriched for processes important for neuromuscular junctions. This suggests possible functions of sXIST in modulating glia–neuron interactions, perhaps via competitive miRNA binding. This idea is also supported by overexpression analysis of a partial sXIST sequence and the finding of significant XIST expression changes in human cardiomyopathy and polyneuropathy. In summary, the current study suggests a novel, non-XCI role of XIST in peripheral Schwann cells that is mediated by a newly recognized transcript.
X-inactive specific transcript (XIST) is a long noncoding RNA (lncRNA) that serves as an important regulator of X Chromosome inactivation (XCI) in mammalian XX females (Loda and Heard 2019). Through interactions between its functional domains (repeat regions A–E) and various proteins, such as Polycomb Repressive Complexes (PRC1/2), CIZ1, and YY1, the XIST ribonucleoprotein complex silences one copy of the X Chromosome in female somatic tissues (Loda and Heard 2019; Jacobson et al. 2022). Since the 1990s, XIST expression has been continuously shown to be female specific and exclusively expressed from the inactive X Chromosome in mammals (Borsani et al. 1991; Brockdorff et al. 1991; Brown et al. 1991). It has also been described to unambiguously target the X Chromosome to silence gene expression (Panning 2008; Cerase et al. 2015). However, recent work indicated that XIST may also have roles in regulating autosomal genes in female cells (Dror et al. 2024; Yao et al. 2025). Moreover, some studies in cancers further reported that XIST could function as a “miRNA sponge” by sequestering multiple miRNAs and thus affecting the expression of their mRNA targets (Marshall et al. 2019; Jiang et al. 2021; Liu et al. 2021; Yan et al. 2022).
Along with these findings of new XIST functions beyond XCI, the canonical view of its exclusive expression in female cells has also been challenged. A few studies have reported rare instances of XIST expression in male cells, mostly in cancers. For example, two groups independently found XIST expression in male samples from multiple types of cancers, such as testicular cancer (Looijenga et al. 1997) and hepatocellular carcinoma (Sadagopan et al. 2022). Similarly, among the 54 human tissues analyzed by bulk RNA sequencing (RNA-seq) in the GTEx consortium from hundreds of healthy individuals (Lonsdale et al. 2013), robust XIST expression was observed in the testes and male tibial nerves (Lonsdale et al. 2013; Sadagopan et al. 2022). Although XIST expression in the testes during spermatogenesis is well documented (Salido et al. 1992; McCarrey et al. 2002), its regulation and potential function in male tibial nerve remain unexplored. In male cancers, however, Sadagopan et al. (2022) suggested that XIST expression could lead to repression of genes on the X Chromosome. Thus, additional studies are needed to address if this occurs in normal human tissues, if repression also occurs for autosomal genes, and, if so, in what cell types and to what extent. XIST RNA may have entirely new tissue- or cell-type-specific roles in males, which further highlights the need for investigation into its noncanonical functions.
Toward this, we performed a systematic analysis of single-cell RNA-seq (scRNA-seq) and chromatin accessibility (scATAC-seq) data to characterize XIST expression in male and female tissues from healthy human individuals, including data from child and adult hearts (Sim et al. 2021; Mehdiabadi et al. 2022; Kanemaru et al. 2023), skeletal muscle (Kedlian et al. 2024), and many other tissues (Zhang et al. 2021). We also analyzed transcriptomic data from multiple studies of mouse peripheral nerves to determine if XIST/Xist expression in male nerves is conserved. We further performed transcriptomic profiling to characterize genes and pathways that may be affected by the expression of a new XIST isoform (termed “sXIST”) that we discovered in male peripheral glia. Using these systematic approaches, we aim to investigate (1) the cell/tissue type and disease relevance of sXIST expression in male cells, (2) distinct transcriptional regulation of the XIST expression in male and female cells, and (3) the potential molecular mechanisms of sXIST functions in peripheral glia. The overarching goal is to understand the potential roles of XIST in peripheral nerves and its relevance to the functions and diseases of peripheral nervous and cardiovascular systems.
Results
XIST expression in human male and female Schwann cells
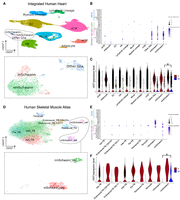
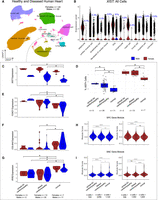
To study the effects of sex chromosomes (XX vs. XY) on gene programs in heart tissues at the single-cell level, we integrated existing scRNA-seq (or single nucleus RNA-seq [snRNA-seq]) data from 20 healthy child and adult human hearts, which were collected from a variety of sources (Supplemental Table S1; Sim et al. 2021; Mehdiabadi et al. 2022; Kanemaru et al. 2023). We unexpectedly observed XIST expression in male hearts. To study this in more detail, we clustered the integrated data to define major cell types in human hearts, from which glial (referred to as “neural” in some previous reports) cells were extracted and further subclustered to identify nonmyelinating and myelinating Schwann cells (Fig. 1) using conventional markers from the literature, such as SCN7A and SLC35F1 for Schwann cells (Karlsson et al. 2021), NRXN1/2 as panglial markers (Diehl et al. 1986), TUBB3 for neurons (Diehl et al. 1986), and PLP1 as a myelination marker (Fig. 1; Supplemental Fig. S1; Diehl et al. 1986). Based on the lack of neuronal marker expression, we determined that the previously annotated “neural” cells were glia, in agreement with the authors of the Heart Cell Atlas (Kanemaru et al. 2023). Our analysis showed that male glia expressed XIST robustly (Fig. 1A–C). To study if this is specific to the heart, we then obtained human skeletal muscle scRNA-seq data for fibroblasts and neural cells (Kedlian et al. 2024), with nonmyelinating and myelinating Schwann cells annotated by the original authors (Fig. 1D–F). In both tissues, male glia showed strong XIST expression (Fig. 1C,F). More specifically, a greater percentage of male nonmyelinating Schwann cells (33.63% in heart, 31.13% in skeletal muscle) expressed XIST compared with their myelinating counterparts (20.29% in heart, 21.71% in skeletal muscle; heart: P-value = 1.7 × 10−10; skeletal muscle: P < 2.2 × 10−16; Fisher's exact test). The average expression of XIST in male nonmyelinating Schwann cells was also higher than in myelinating cells, determined at either the single-cell level (heart: adjusted P-value = 2.4 × 10−7, AvgLog2 fold-change [FC] = 1.2; skeletal muscle: adjusted P-value = 3.3 × 10−33, AvgLog2FC = 8.9; Wilcoxon rank-sum test) (Fig. 1C,F) or pseudobulk level with cells from the same samples pooled (heart: adjusted P-value = 2.1 × 10−6, log2FC = 2.8; skeletal muscle: adjusted P-value = 2.8 × 19−8, log2FC = 5.6; negative binomial test). In general, a greater percentage of male glia expressed XIST compared with other cell types (<0.55%) in both data sets (Supplemental Table S2; Supplemental Fig. S2). Additionally, even among cell types with XIST RNA detected, XIST abundance remained significantly lower in the male glia than in female glia (Fig. 1C,F).
High XIST expression in nonmyelinating Schwann cells from male human heart and skeletal muscle tissue. (A) Clusters of 510,171 human heart cells from 11 male and nine female individuals. Cells within the dashed box are glia, which are colored by subtypes. (EC) Endothelial cells, (FB) fibroblasts, (aCM) atrial cardiomyocytes, (vCM) ventricular cardiomyocytes, (mSchwann) myelinating Schwann, (nmSchwann) nonmyelinating Schwann. (B,E) A variety of positive and negative markers used to identify male and female myelinating and nonmyelinating Schwann cells. (C,F) Seurat-normalized XIST expression across all cells, grouped by cell types and split by sexes. The asterisk represents a significant difference in expression between male nonmyelinating (nm) and myelinating (m) Schwann cells. (D) Clusters of 20,611 skeletal muscle fibroblast and neural cells from eight male and four female individuals, colored by cell annotations provided by the original authors.
To expand our finding, we analyzed XIST expression in the Human Cell Atlas using the CZ CELLxGENE discover gene expression tool (CZI Cell Science Program et al. 2024), which enabled us to quantify the percentages of cells expressing XIST across a large number of cell types in aggregated data sets from multiple tissues. Among all the cell types in the atlas, we found significant XIST expression in male Schwann cells (58% of 2955 cells) and nonpigmented ciliary epithelial cells (54% of 3011 cells) of the eye, pancreatic stem cells (59% of 492 cells), and fibro/adipogenic progenitor cells of adipose tissue (51% of 1154 cells) (Supplemental Table S3). Although XIST expression was sporadically detected in other male cell types, these were the only cell types with expression in >50% of cells, which accounted for just 0.15% (5/3151) of all cell type–tissue combinations that we extracted from the database up to January 2025. There were still only 0.98% (31/3151) of all cell type–tissue combinations if we reduced the detection percentage to >30%, which was approximately the percentage of male cells expressing XIST in the heart and skeletal muscle data (Fig. 1C,F). In the central nervous system, 6% of male differentiation-committed oligodendrocyte precursors in both the spinal cord and brain expressed XIST, but only 0.4% of mature oligodendrocytes in the spinal cord and 0.3% in the brain expressed XIST. Male XIST expression was barely detectable in two large cohorts of human brains (Velmeshev et al. 2019; Wamsley et al. 2024).
Taken together, our analysis found that XIST is consistently expressed in male Schwann cells, whereas its male expression in other cell types is much sparser and probably needs stronger evidence from more studies (Supplemental Fig. S3; Supplemental Table S3). Note that our finding of XIST expression in male cardiac glia was independently described by a recent report from Gorin and Goodman (2025).
Characterization of XIST transcript sequence and regulation in peripheral nerves
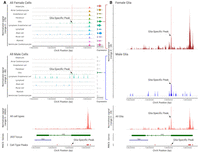
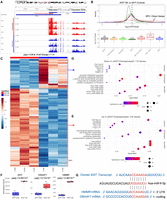
Next, we sought to investigate how XIST achieves such specific expression by comparing chromatin accessibility data from assay for transposase-accessible chromatin using sequencing (ATAC-seq) across cell types. We first turned to scATAC-seq data from the Human Heart Cell Atlas, which were generated using the 10x Multiome platform (Kanemaru et al. 2023). We recalled ATAC-seq peaks using MACS2 (Zhang et al. 2008) for each of the broad cell types defined by the authors, except we renamed their “neural” cells as “glia” because they lacked expression of the neuronal markers in Figure 1. This identified two ATAC peaks at the XIST locus, with a peak called only for the cardiac glia ∼10 kb downstream from the main XIST promoter. The main XIST promoter is constitutively active in female cells but not in male cells (Fig. 2A). The glia-specific peak (Chr X: 73,841,364–73,841,611; hg38) is located in a region suggested to be an active promoter in several cell lines by the FANTOM5 project. Their data are from a combination of cell lines and tissues (including Schwann cells, the optic nerve, and skeletal muscle), with the sample metadata indicating 32% males, 32% females, and 36% as other/NA (Supplemental Fig. S4A; Noguchi et al. 2017). Additionally, the binding sites for a variety of transcription factors (TFs), such as FOXD3 and REST, are found in this region according to motifs in the JASPAR database (Supplemental Fig. S4B; Supplemental Table S4; Rauluseviciute et al. 2024). Among the predicted TFs, one (FOXO1) was expressed higher in both skeletal muscle and heart nonmyelinating Schwann cells compared with myelinating, whereas 11 (NFATC3, SOX13, PBX1, RREB1, SOX10, TCF4, ETS1, SREBF1, CEBPB, KLF2, and ETV5) were higher in either skeletal muscle or heart nonmyelinating Schwann cells. Analysis of the transcription regulatory programs by the SCENIC software predicted that FOXO1 (and YY1) could regulate XIST in heart and skeletal muscle Schwann cells, with greater activity in nonmyelinating cells (Supplemental Fig. S5). This suggests that FOXO1, among other TFs, may regulate a previously uncharacterized promoter (or enhancer) of XIST.
Chromatin accessibility at the XIST locus across human heart cell types. (A) Multiomic scATAC/RNA-seq signal and expression (split by sex) across cell types. MACS2-called peaks are represented as “cell-type peaks.” The normalized signal across all cells and fragments for 100 random cells are presented above the XIST gene locus, with introns and exons as thin and thick green lines, respectively. (B) A zoomed-in view of scATAC-seq signal for male (blue) and female (brown) glia. (A,B) The red highlighted region represents the glia-specific peak (Chr X: 73,841,364–73,841,611) called by MACS2.
After separating male from female cells, we observed that all female cell types show high chromatin accessibility at the main XIST promoter, consistent with constitutive XIST expression in females; however, it became clear that the alternative promoter is likely active in both males and females. Silencing of the main XIST promoter in males allowed us to correlate the increased accessibility of the alternative promoter with increased XIST expression (Fig. 2A,B). There were significantly more scATAC fragment counts detected over the alternate promoter region (Chr X: 73,841,364–73,841,611 ± 100 bases) in glia compared with all other cell types for both males (P-value = 2.83 × 10−14, AvgLog2FC = 3.98; Wilcoxon rank-sum) and females (P-value = 1.92 × 10−11, AvgLog2FC = 2.84; Wilcoxon rank-sum). This was associated with significantly increased XIST expression in male glia (P-value < 2.23 × 10−308, AvgLog2FC = 10.51; Wilcoxon rank-sum test) and to a lesser degree in female glia (P-value = 1.24 × 10−4, AvgLog2FC = 0.37) compared with all other cell types, with the latter likely masked by the higher expression of the full-length XIST in female cells.
To study if the newly discovered alternative promoter leads to alternative XIST transcripts, we analyzed bulk RNA-seq data because most of the scRNA-seq technologies sequence only the 3′ end of RNAs. Unfortunately, we were unable to find bulk RNA-seq data for purified male Schwann cells. Thus, we turned to publicly available bulk RNA-seq data from peripheral nerve samples because this tissue type contains a relatively high proportion of Schwann cells, which are functionally required for supporting/myelinating peripheral neurons. In a previous study, Sandy-Hindmarch et al. (2025) performed strand-specific bulk RNA-seq on Morton's neuroma and control samples from males and females independently. These samples were collected from a variety of digital, upper limb, and lower limb nerves, allowing us to examine the distribution of RNA-seq reads along the XIST locus in both strands. The data showed that reads from male samples were significantly enriched at the last exon (i.e., exon 6 at the 3′ of XIST) (Supplemental Fig. S6). Although RNA-seq reads were also mapped to other exons (exons 2–5), only a few reads were aligned to the end of the first exon in males. A comparison of the ratios of RNA-seq reads mapped to the first and sixth exons in male and female samples support a significant bias of reads to the latter in males (P-value = 0.044; t-test) (Supplemental Fig. S6A). Importantly, the region at the end of the first exon with only a few RNA-seq reads in male samples is near the cardiac glia–specific accessible region discussed above (Fig. 2).
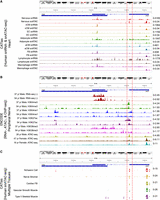
To seek additional, independent support for the activation of the alternative promoter, we next utilized the cis-element atlas database (CATlas) (Zhang et al. 2021) to study chromatin accessibility for other cell types at the XIST locus because it curates comprehensive reference maps of cis-regulatory elements from thousands of cell types. Moreover, it has a heart data set with both snRNA-seq data and scATAC-seq peaks from males and females. We found that cells annotated as “nerve” showed increased chromatin accessibility at the alternative XIST promoter, with concordant snRNA-seq reads in the same region, but other cell types did not (Fig. 3A).
Assessment of ATAC-, RNA-, and ChIP-seq read coverages over the XIST locus. (A) snRNA/ATAC-seq tracks from CATlas heart data set. (B) RNA-seq (±strands) and H3K4me3, H3K4me1, H3K27ac, and H3K36me3 ChIP-seq from two human male tibial nerve samples and ATAC-seq tracks from one human male and two female sciatic nerve samples in the ENCODE database. (C) snATAC-seq tracks from CATlas multitissue data set. (A–C) All plots show the region at Chr X: 73,790,905–73,854,216 from the hg38 reference genome. The 3′ region of XIST exon 1 is enclosed by two red dashed lines. TSIX gene location spanning Chr X: 73,792,205–73,829,231 on the + strand is shown as a blue bar, indicating the RNA-seq reads were from XIST but not TSIX. For each track, cell types and/or sequencing modality are shown on the left, and the data range is shown in the right. Additional data for the tibial and sciatic nerve samples are in Supplemental Figure S7.
We also analyzed tibial nerve bulk RNA-seq and ChIP-seq data (two male samples) and sciatic nerve ATAC-seq data (one male sample) available from the ENCODE Project (The ENCODE Project Consortium 2012; Luo et al. 2020). The results support open chromatin accessibility at the alternative promoter, with its activity further supported by active histone modifications H3K4me3 (marks promoters) and H3K27ac (marks both promoters and enhancers). Moreover, the data strongly support that XIST expression in males occurred downstream to the alternative promoter, as seen from enriched signals of RNA-seq and H3K36me3, a histone mark for transcription elongation (Fig. 3B). Like in the above analysis of Morton's neuroma data, we again found that RNA-seq reads exhibited a significantly greater bias to exon 6 (vs. exon 1) in the male nerve samples compared with the male brain samples (P-value = 0.0011; t-test), which do not have Schwann cells (Supplemental Fig. S7). This biased trend was also in female data, but the difference was not statistically significant (P-value = 0.22).
To address the concern of inferring RNA transcript sequences from short-read technology, we searched the ENCODE database for XIST transcripts from long-read sequencing technology. There was no such data from nerve samples, but we found RNA transcripts that aligned with the presence of sXIST transcripts in human hearts (Supplemental Fig. S8, red arrows, a male and a female sample).
Lastly, for additional regulation evidence, we examined another CATlas data set derived from integration of multiple male and female tissues and found that adult Schwann cells contained an ATAC-seq peak (called by the authors) at Chr X: 73,841,318–73,841,717, which was essentially the exact same region where we called an ATAC-seq peak for cardiac glia (Fig. 2). The same peak was called for cardiac fibroblasts, peripheral nerve stromal cells, vascular smooth muscle cells, and type II skeletal myocytes in CATlas (Fig. 3C), potentially related to sparse XIST expression in nonglial heart cells (Fig. 1). Nevertheless, the peaks were the highest in Schwann and peripheral nerve stromal cells. Note that no peak at this region was called for the remaining 106 adult cell types or any of the 111 fetal cell types in CATlas.
Taken together, our systematic analysis uncovers multiple lines of evidence that an alternative XIST promoter is active in peripheral glia, most likely Schwann cells, resulting in a shorter XIST transcript (or transcripts) in both males and females, which we referred to as sXIST.
Functional implications of XIST activation in male Schwann cells/nerves
To study possible roles of sXIST expression, we first addressed whether it is involved in repressing X Chromosome genes in males. We divided the genes on the X Chromosome into groups: genes undergoing X inactivation (X nonescapees), genes escaping inactivation (X escapees) (Wainer Katsir and Linial 2019), and genes in the pseudoautosomal region (PAR) (Weng et al. 2016). As controls, we also included Y Chromosome genes that are not in the pseudoautosomal region (Y non-PAR) and all autosomal genes. We compared their expression between myelinating and nonmyelinating Schwann cells using DESeq2 (Love et al. 2014) and the pseudobulking method, dividing samples by sex (Fig. 4A,B).
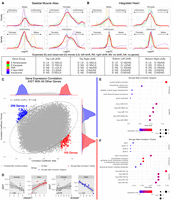
Potential roles of XIST in peripheral nerve. (A,B) Density plots of log2FC from a variety of gene group comparisons using human skeletal muscle data (A) and human heart data (B) along with their expected and observed trends. For X Chromosome nonescapee genes, the expected trend is based on the presumption that sXIST participates in XCI. In all comparisons, the log2FC values were computed from log2(sample-on-right/sample-on-left). Gene groups with too few genes for robust density analysis in a comparison were not included in these plots. Expected and observed trends for A and B are summarized at the bottom. (C) Pearson correlation coefficients (r) between XIST and all other genes computed separately for males (x-axis) and females (y-axis) using peripheral nerve bulk RNA-seq data. In this plot, each dot is a gene, and the predicted 99% confidence ellipse was overlayed (refer to Supplemental Fig. S12). Outlier genes with a male Pearson's correlation P-value < 0.05 and absolute difference >0.3 between male and female correlation coefficients were colored red if male r > 0 and blue if male r < 0. (D) Two gene examples (DNAAF1 and KCNAB1) exhibiting stronger positive (red) and negative (blue) expression correlation with XIST in males. Expression values are provided as log2(DESeq2-normalized counts + 1). (E,F) Overrepresentation analysis for genes with stronger negative male correlations (E) or stronger positive male correlations (F). Dot size represents the number of query genes in each term, and color is −log(FDR-adjusted P-value).
If sXIST participates in X Chromosome silencing in males as it does in females, we would expect a general decrease in the expression of X nonescapee genes in male nonmyelinating (higher sXIST) compared with myelinating Schwann cells, manifesting as a leftward shift in the distributions of their log2FC values in Figure 4, A and B. No global shift was observed in the comparisons of skeletal muscle cells (Fig. 4A). Although the mean log2FC value of this comparison in the heart data was −0.81 with a slight leftward shift (i.e., most nonescapee genes reduced expression in nmSchwann cells), this trend was also seen in our control groups (autosomal genes and X-escapee genes) and similarly in the comparisons of female samples. Thus, it is unlikely that sXIST leads to selective repression of X Chromosome genes. Because male cells have only one X Chromosome, we also expected a decrease in X nonescapee expression in males (leftward shift) when comparing male to female nmSchwann cells. Instead, the distribution of log2FC values were centered around zero without apparent skewness for both human and skeletal muscle data sets. The same trends were observed when male and female mSchwann cells were compared. The expected and observed trends are summarized at the bottom of Figure 4, A and B, and the mean log2FC values for each comparison can be found in Supplemental Figure S9. Furthermore, no evidence for a global reduction of nonescapee genes was observed in male or female Schwann cells compared with all the other cell types (Supplemental Fig. S10).
Only PAR genes showed a consistent expression reduction in nmSchwann relative to mSchwann cells in males for both heart and skeletal muscle data sets (shown as left-shift in Fig. 4A,B, top), indicating a global decrease in PAR gene expression in nonmyelinating Schwann cells (heart: mean log2FC = −0.87; skeletal muscle: mean log2FC = −0.27). PAR genes are thought to escape XCI (Helena Mangs and Morris 2007), but we found that they exhibited higher expression in males compared with females irrespective of Schwann cell type (Fig. 4A,B, bottom). This trend appears the same in other tissues from our analysis of bulk GTEx data (Supplemental Fig. S11; Lonsdale et al. 2013). The increased PAR expression in males is consistent with previous reports (Tukiainen et al. 2017). Thus, the increased PAR expression in male compared with female Schwann cells can be attributed to sex biases, whereas decreased PAR gene expression in nonmyelinating versus myelinating Schwann cells (in males) may be cell type dependent and related to sXIST expression.
In short, we did not find convincing evidence for the involvement of sXIST in the general suppression of X Chromosome genes in the male nonmyelinating Schwann cells. This contrasts with previous reports in male cancer cells (Looijenga et al. 1997; Sadagopan et al. 2022). Nevertheless, our results are consistent with the consensus view in the literature that XCI needs the functional domains in XIST exon 1 (Brockdorff 2002), which are absent in sXIST.
Next, we reasoned that genes potentially affected or regulated by sXIST would exhibit a stronger correlation with XIST expression in male cells than in female cells because the correlation in female cells captures the combined expression of sXIST and the full XIST transcript, with the latter at a much higher level, thus masking sXIST effects in this analysis. To investigate this, we used the Morton's neuroma bulk RNA-seq data set from peripheral nerves (Sandy-Hindmarch et al. 2025) discussed above (Supplemental Fig. S6) because scRNA-seq data are too sparse for this purpose. We first randomly selected 100 genes and calculated the Pearson correlation coefficients between each of them and all the other human genes across all the male (n = 11) and female (n = 22) samples separately to obtain a null distribution defined by fitting an ellipse around 99% of the data points (Supplemental Fig. S12A,B). Adding XIST, we found that 2.93% of all genes correlated with XIST were outside the ellipse (i.e., outliers), compared with just 0.81% in null background (P-value < 2.2 × 10−16, odds ratio [OR] = 3.68; Fisher's exact test), thus representing genes with significantly different correlations with XIST between male and female samples. In the end, we determined that 195 genes (0.76% of all genes) had a stronger and positive correlation (SPC) with XIST in males than in females compared with 0.17% of control gene pairs (P-value < 2.2 × 10−16, OR = 4.61; Fisher's exact test), and 206 genes (0.80% of all genes) had a stronger and negative correlation (SNC) with XIST in males, compared with 0.16% of control comparisons (P-value < 2.2 × 10−16, OR = 4.92; Fisher's exact test) (Fig. 4C). Two examples, DNAAF1 (SPC) and KCNAB1 (SNC), are shown in Figure 4D.
Among the 206 SNC genes, many have roles in neuromuscular system development and function, including CYFIP2 (involved in synapse formation and plasticity), FLYWCH1 (linked to neuronal migration and axon guidance), ion channel genes (e.g., SLC2A4, KCNQ4, KCNAB1, and KCNMB1), calcium signaling genes (CACNA1S, CACNA1C, and CACNB2), extracellular matrix genes (COL4A2 and ITGA8), and genes related to muscle structure and function (ACTA1/2 and ACTG2). Overrepresentation analysis (using ToppFun) (Chen et al. 2009) of the SNC genes also revealed a significant enrichment (adjusted P-value < 0.05) for functions related to muscle system process, calcium ion homeostasis, skeletal muscle development, actin filament–based movement, and action potential (Fig. 4E). Among the 195 SPC genes, many have functions in neurodevelopment and synaptic function (e.g., FOXP2, NPAS1, and CNTNAP4), signaling transduction (e.g., NFATC1, WNT9A, and RASGRF2), ciliary function (e.g., DNAH9 and DNAAF1), and immune response (e.g., CXCR4 and IL18). Unbiased enrichment analysis of the SPC genes showed a significant enrichment of functions related to cilium, extracellular transport, and axoneme assembly (Fig. 4F). Although research in this area is relatively sparse, primary cilia are signaling hubs, exist in glia (and neurons), exhibit impairment in some neurodegenerative diseases, and may be involved in a variety of functions related to nerve regeneration signaling in Schwann cells (Ki et al. 2021). Note that neither SPC nor SNC genes were enriched on the X Chromosome, and very few exhibited a significant expression difference between male and female samples. Interestingly, both SPC and SNC genes were enriched for putative targets of multiple miRNAs (Fig. 4E,F). Previous studies have reported that XIST can act as a molecular sponge to inhibit miRNA function (Marshall et al. 2019; Jiang et al. 2021; Liu et al. 2021; Yan et al. 2022), which will lead to a positive correlation among genes sharing miRNA targets because of fewer miRNAs binding to individual targets (Hausser and Zavolan 2014). Among the top 15 miRNAs predicted to target SPC genes, five of them (miR-361-3p, miR-32, miR-149-3p, miR-122, and miR-367) have reported evidence for interactions with XIST (Li et al. 2018; Cheng et al. 2020; Yang et al. 2020; Jiang et al. 2021; Wang et al. 2023).
To relate this result back to Schwann cells, we wondered how the SPC and SNC genes were differentially expressed between nonmyelinating and myelinating Schwann cells. First, we found that two SPC genes (RNF144A and PCBP3) were significantly upregulated (adjusted P-value < 0.05 from DESeq2) when comparing all heart nonmyelinating to myelinating Schwann cells. For skeletal muscle, three SPC genes (CCND3, ANO4, and PYGL) were upregulated. For SNC genes, heart nonmyelinating Schwann cells showed significant downregulation of 31 (e.g., ILK, FLNA, and CYFIP2), whereas no SNC genes were downregulated in skeletal muscle. We next used biomaRt to extract all genes in the “epithelial cilium movement involved in extracellular fluid movement” (GO:0003351, 43 genes) and “actin filament–based movement” (GO:0030048, 133 genes) categories because SPC and SNC genes were enriched for these terms, respectively. We used these genes to compute module scores for nonmyelinating, myelinating, and “other” glia in the integrated human heart scRNA-seq data discussed above (but not skeletal muscle owing to the relatively low number of total Schwann cells). The module scores using the cilia-related gene set were significantly higher in nonmyelinating Schwann cells compared with myelinating and “other” for both males and females (P-value < 0.05; Wilcoxon rank-sum) (Supplemental Fig. S12C). However, the module scores using the “actin filament–based movement” gene set were not consistently lower in nonmyelinating Schwann cells compared with myelinating (Supplemental Fig. S12D).
Overall, these results indicate that XIST-expressing male nonmyelinating Schwann cells do not exhibit evidence of XCI; instead, they suggest sXIST may have other functions potentially involving miRNAs, including some targeting cilia-related genes.
Assessing the effects of sXIST overexpression in a human cell line
To further assess if sXIST has non-XCI functions, we overexpressed a subsequence of sXIST in a human cell line (HEK-293T), more specifically the second half of XIST exon 6 (Supplemental Methods; Supplemental Fig. S13). The construct lacks domain E, which is required for anchoring XIST RNA to the inactive X Chromosome through CIZ1 interactions (Sunwoo et al. 2017), and other domains needed for XCI. We carried out strand-specific RNA-seq analysis comparing sXIST overexpression (XIST OE) to GFP controls (n = 3; Fig. 5) and identified a few hundred differentially expressed genes (DEGs), including XIST itself (Fig. 5A,F; Supplemental Table S5). Among the various gene groups described in Figure 4, the SPC genes and, especially, the 10 genes in cilia-related pathways exhibited strong, increased expression in the XIST OE samples (Fig. 5B). However, the other groups of genes did not exhibit a group-level trend, except for a reduction of PAR genes (Fig. 5B; Supplemental Fig. S13C). Thus, our overexpression data are consistent with our bioinformatic results in Figure 4.
RNA-seq analysis of sXIST overexpression. (A) IGV tracks showing RNA-seq reads over Chr X: 73,790,905–73,854,216, with TSIX and XIST indicated. Samples overexpressing the second half of exon 6 (XIST OE) are shown in red, and GFP controls (Ctrl) are in blue. (B) Density and boxplots showing log2FC values across gene groups. Values above boxplots are the mean log2FC for that group. (C) Heatmap of the DEGs (adjusted P-values < 0.05 and |fold change| > 1.5). (D,E) ToppFun results from downregulated (D) and upregulated (E) genes in C. (F) Boxplots showing the expression of XIST, DNAAF1, and HMMR. (G) Cloned XIST, DNAAF1, and HMMR transcript sequence alignments with hsa-miR-9-5p.
Functional enrichment analysis of the 318 significantly upregulated (adjusted P-value < 0.05 and fold change > 1.5) and 119 downregulated (adjusted P-value < 0.05 and fold change < −1.5) (Fig. 5C,D) genes showed that the upregulated set was significantly enriched for GO terms related to cilia and axoneme assembly (Fig. 5E). Moreover, “cilium movement” and “axoneme assembly” were in the top five most enriched GO terms for both male SPC and XIST OE upregulated genes. Four upregulated genes (BDKRB2, DNAAF1, DNAI3, and LRRC46) and two downregulated genes (COL4A6 and P2RX1) in our overexpression analysis overlapped with SPC and SNC genes, respectively. The small overlap potentially reflects distinct biological contexts of HEK-293T cells and peripheral nerves.
Targets of multiple miRNAs were enriched in the upregulated genes. Importantly, among the top 20 miRNAs, miR-9, miR-651, miR-520a-5p, and miR-218-2 (also called miR-218-3p) were predicted by miRDB (Chen and Wang 2020) to bind to the overexpressed sXIST sequence, as was miR-520a-3p, but it was not ranked in the top 20. miR-9-5p is especially interesting because it is expressed in both HEK-293T (Kavakiotis et al. 2022) and nerve tissue (Rishik et al. 2025), previously implicated in XIST sponging (Zheng et al. 2020), and inhibits Schwann cell migration (Zhou et al. 2014). One of its predicted targets is hyaluronan-mediated motility receptor (HMMR) (Fig. 5F,G), which is highly expressed in Schwann cells (Ouasti et al. 2020), is important for their motility (Ouasti et al. 2020), plays a role in mitotic spindle positioning (Connell et al. 2017), and supports neural development (Connell et al. 2017). Although not predicted to bind to its 3′ UTR, a potential hsa-miR-9-5p binding site is found in the final exon of DNAAF1 (Fig. 5G), a cilia-related gene strongly correlated with XIST expression in males (Fig. 4E). It has been implicated in neural tube defects (Miao et al. 2016) and ciliary dyskinesia owing to its important role in dynein-arm complex assembly (Loges et al. 2009).
As for the other miRNAs, cilia-related genes are predicted targets of miR-520a-5p (DNAH6, DNAI4), miR-651 (DNAAF1), miR-218-2 (DNAAF1), and miR-520a-3p (DNAAF1). Mir-218-2 and miR-34a-5p were also predicted to target LRRC46. Additionally, miR-520a-5p, miR-9, and miR-34a have been previously implicated in XIST sponging (Liu et al. 2018, 2021; Zheng et al. 2020). To check if these miRNAs are expressed in HEK-293T cells and nerve tissue, we searched the microRNA Tissue Expression Database (Kavakiotis et al. 2022) and confirmed that miR-218-2-3p, miR-34a-3p/5p, miR-651-3p/5p, and miR-9-3p/5p were expressed, with miR-9 showing the strongest expression (top 3% of 1773 expressed miRNAs in 293T cells and top 1% of 1089 expressed miRNAs in peripheral nerves). These miRNAs were also detected in nerve tissue according to the miRNA Tissue Atlas (Rishik et al. 2025).
Taken together, many genes, including those related to cilia, were upregulated upon overexpression of an sXIST fragment, and there is no evidence that the DEGs are enriched on the X Chromosome. The overexpression data, however, support the potential function of sXIST as a miRNA sponge.
Evaluating XIST expression in glial cells from human cardiomyopathy and polyneuropathy tissue
Because sXIST exhibited high expression in male Schwann cells and because Schwann cells are part of the intracardiac nervous system and are important for cardiac sympathetic nerve fasciculation (i.e., axon bundling to form nerve tracts) (Hortells et al. 2021), we wondered whether there might be changes in XIST expression in the glia of diseased hearts, more specifically from patients with arrhythmogenic cardiomyopathy and dilated cardiomyopathy (up to 38% of hearts with dilated cardiomyopathy exhibit nonsustained ventricular arrhythmias) (Spezzacatene et al. 2015). We thus analyzed a snRNA-seq data set with 881,081 nuclei from 18 healthy and 61 nonischemic, failing hearts (Fig. 6A; Reichart et al. 2022). We again found that author-annotated male “neural” cells, which we reclassified to glia based on marker gene expression (Supplemental Fig. S14), expressed XIST at much higher levels compared with other male cell types (adjusted P-value < 2.23 × 10−308, log2FC = 6.44; Wilcoxon rank-sum) (Fig. 6B). We also found that the percentage of cells expressing XIST in glia was significantly higher in both cardiomyopathy groups for males and females (P-value = 2.2 × 10−16, OR = 1.14; Fisher's exact test) (Fig. 6D). After pseudobulking by samples, the expression difference of XIST between disease groups and controls was only significant for males (Fig. 6C). High expression of the full-length XIST transcript in female samples could explain the inconsistency in the results from the pseudobulk level and the single-cell level, because sXIST would contribute to a fraction of the total pseudobulked expression values.
XIST expression in arrhythmogenic and dilated cardiomyopathy. (A) UMAP of 881,081 heart nuclei from snRNA-seq data performed on 61 heart failure patients and 18 healthy controls (Reichart et al. 2022). Cells originally labeled “neural” were renamed to “glia” based on expression of glial markers (Supplemental Fig. S14). (B) Normalized XIST expression across all nuclei, grouped by cell type and split by sex. For males, the percentages of cells expressing XIST are shown at the top. (C–I) Cells were subset by glia cell–type annotation and grouped based on arrhythmogenic (arrhyth) RV cardiomyopathy, dilated cardiomyopathy, and normal (healthy) annotations and sex. Normalized XIST (C), FOXO1 (E), COL4A6 (F), and RYR2 (G) expression after pseudobulking glial cells by donor. (D) The percentage of glial cells that express XIST for each sample by disease group and sex. (H,I) Module scoring was performed on SPC (E) and SNC (I) genes from Figure 4. Asterisks indicate a significant difference using Fisher's exact test (D) or DESeq2 negative binomial test (C,E–I).
Because of XIST upregulation in cardiomyopathy, we expected that SPC genes would be upregulated and SNC genes downregulated (Fig. 4) in both types of cardiomyopathies. We found this to be true for five genes, which were RYR2, TPM2, CACNB2, AKAP6, and MYOCD. A full list of the overlap between disease DEGs by SPC/SNC genes, sex, and expression direction relative to normal hearts is provided in Supplemental Table S6. We also observed significant differences in FOXO1 expression, mentioned above as a potential TF regulating sXIST expression (FOXO1 expression was significantly higher in male and female disease group glia, except female arrhythmogenic RV cardiomyopathy) (Fig. 6E). COL4A6, which is an SNC gene (Fig. 4) and downregulated in the XIST OE group (Fig. 5), showed decreased expression in male and female dilated cardiomyopathy glia compared with healthy hearts (Fig. 6F). RYR2, another SNC gene that encodes a receptor crucial for cardiac muscle function and is involved in cardiac and neuronal disorders (ryanopathies) (Sleiman et al. 2021), was significantly downregulated in both types of cardiomyopathies for male and female glia (Fig. 6G).
For males and females, we also used SPC and SNC genes to compute module scores for glia from cardiomyopathy and healthy groups. For males, the SPC module scores, as expected, were significantly higher (P-value < 0.05; Wilcoxon rank-sum test) in glial cells from both types of cardiomyopathies compared with controls (Fig. 6H). For females, module scores were higher for glial cells from arrhythmogenic RV cardiomyopathy but not dilated cardiomyopathy (Fig. 6H). For both males and females, SNC gene module scores were significantly lower in glial cells from both cardiomyopathies compared with healthy controls (Fig. 6I).
Likewise, we studied XIST and its putatively affected pathways using scRNA-seq data comparing polyneuropathy (PNP) sural nerve samples to controls, which included nonmyelinating, myelinating, and repair Schwann cells (Heming et al. 2025). We found that XIST was significantly downregulated in nonmyelinating Schwann cells from PNP samples (adjusted P-value = 2.98 × 10−2, AvgLogFC = −1.91;two-sided likelihood ratio test) but not in myelinating or repair Schwann cells (Supplemental Fig. S15; Supplemental Table S7). Heming et al. (2025) noted that genes upregulated in nonmyelinating Schwann cells from PNP samples were associated with cell migration. We further found that the average logFC value for genes in the actin filament–based movement (GO:0030048) gene set was 0.56 (high in PNP) comparing PNP nonmyelinating Schwann cells versus controls, whereas the average logFC values for cilium-related genes (GO:0003351) was −0.19 (lower in PNP).
Overall, our results indicate that peripheral glia from diseased tissue exhibit significant changes in XIST expression, but the direction of change depends on the disease (lower in PNP and higher in cardiomyopathy), suggesting that XIST (specifically sXIST because it is the only apparent transcript in male hearts) could potentially affect the expression of some genes implicated in heart disease and neuropathy.
Xist expression in mouse male glial cells
To determine if Xist expression is conserved in male mouse Schwann cells, we focused on mouse scRNA-seq data from peripheral nerves (Gerber et al. 2021; Yim et al. 2022). The data were collected after male (n = 2) and female (n = 2) samples were pooled; thus, sex information was not available for individual cells. We therefore used Y Chromosome gene expression to predict sex, similar to the method described by Twa et al. (2025). We found that labeling cells with any nonpseudoautosomal Y Chromosome gene expression as from males was 91.18% accurate using human skeletal muscle cells and 75.95% accurate using human heart cells (Supplemental Fig. S16), but our method is expected to put many true male cells into the female group. We applied this strategy to separate male cells in the scRNA-seq data collected by Gerber et al. for peripheral nerves at postnatal day 1 (P1) and 60 (P60). The authors had identified Schwann cells, so we retrieved these cells from their full data and then subclustered. A cell type in P1 called fibroblast-related (FbRel) also expressed Schwann cell–related genes and were therefore included. For both P1 and P60 data, we used the same myelin-related genes in Figure 1 (PLP1, MPZ, MBP, and PRX) to separate cells into myelinating versus nonmyelinating cells. It is important to note that all clusters expressed myelinating genes to some degree, but those with lower expression were called nonmyelinating Schwann cells (Supplemental Fig. S17A,B). The second data set from Yim et al. already had myelinating and nonmyelinating Schwann cell subtypes separated, so we directly compared cells in these two subtypes after predicting male and female cells (Supplemental Fig. S17C). We should also note that the P60 sciatic nerve sample had only 414 Schwann cells compared with 3443 at P1 and 9676 for other peripheral nerves.
The predicted male Schwann cells from both studies expressed Xist, indicating coexpression of Xist and Y Chromosome gene(s) in the same cells. Nonmyelinating Schwann cells had the greatest percentage of male cells expressing Xist in all three groups of samples (Supplemental Figs. S18, S19). Yim et al. data exhibited greater between-sample variation in the percentage of cells expressing Xist by cell type for both males and females (Supplemental Table S8; Supplemental Fig. S19). A summary of these comparisons for each sample is in Supplemental Table S9, which supports higher Xist expression in the nonmyelinating compared with myelinating Schwann cells (Supplemental Fig. S18A–C), consistent with the human data (Fig. 1). Gerber et al. also provided a tool for their scRNA-seq sciatic nerve atlas (SNAT) data for visualizing gene expression through mouse age and pseudotime by cell type in their Smart-seq2 data. Although Xist was not detected in the Smart-seq2 samples (perhaps a technical effect), Foxo1 (predicted by SCENIC to regulate XIST in humans) exhibited higher expression in nonmyelinating glia, showed greater expression with increasing mouse age, and demonstrated increasing expression through pseudotime for nonmyelinating glia but not myelinating glia (Supplemental Fig. S20). Foxo1 expression was also higher in predicted male and female nonmyelinating Schwann cells in the mouse data we analyzed (Supplemental Fig. S18).
To expand our analysis to other mouse tissues, we again utilized the CZ CELLxGENE discover gene expression tool (CZI Cell Science Program et al. 2024) to determine the percentage of male and female cells expressing Xist in mice by cell type across tissues (Supplemental Fig. S3B). Across all tissues, cells in the eye had a particularly high percentage of male cells expressing Xist, especially for amacrine cells (45%) and retinal ganglion cells (34%). Several other amacrine cell subtypes also had a high percentage of male cells expressing Xist as well as “neural” cells of the eye (7%), which may be glia. A summary of CELLxGene percentages by cell type for mice can be found in Supplemental Table S10.
Taken together, our analysis of mouse RNA-seq data indicate that, like humans, male Schwann cells express Xist with a bias toward the nonmyelinating lineage. The Xist expression in mouse male peripheral glial cells, however, seems less specific than in human data. At this point, it is unclear if this represents a true species difference or a bias in sampling.
Discussion
XIST is the most-studied lncRNA, whose main and canonical functions, interacting proteins, and individual functional domains have been dissected by many previously published works. In this study, we found that a shorter XIST isoform is expressed in male Schwann cells in both healthy and disease conditions, with its level higher in nonmyelinating Schwann cells. Our findings challenge the idea that XIST is exclusively female specific in noncancerous cells, whereas our results support potential non-XCI roles of XIST in both male and female peripheral glia cells.
Our data indicate that XIST expression in male Schwann cells is driven by a previously uncharacterized promoter at the end of the first exon, leading to a shorter XIST RNA. This is based on multiple lines of evidence, including RNA-seq read coverage, H3K36me3 modification, and long-read full-length transcripts. We believe this short XIST isoform is also expressed in females based on shared scATAC-seq peaks across data sets and long-read RNA-seq data from the heart; however, high expression of the full-length isoform makes it impossible to conclude unambiguously if the same short transcript is present in both male and female glia. Furthermore, although our presentation considered one short isoform, sXIST, our data do not exclude the possibility that multiple short, spliced isoforms exist in the Schwann cells because our current data did not have the resolution to resolve alternative splicing. Distinct XIST isoforms have been reported (Johnston et al. 1998; Memili et al. 2001; Yue and Ogawa 2018), but the documented isoforms are not related to the sXIST reported here. We should also mention there is some degree of sXIST expression variations across human individuals from whom all the bulk or single-cell data were derived. In addition to technical reasons, one possibility is that the tissue samples could have different composition of nonmyelinating Schwann cells or that, moreover, sXIST expression is affected by the activation state of Schwann cells.
Our finding brings up more questions than we could address here. The central question is if sXIST has a non-XCI functional role in Schwann cells and, more generally, in peripheral nerves or if its expression is merely transcriptional noise. Because the sXIST transcript(s) does not contain the tandem repeat domains required for XCI, we think its expression does not lead to XCI, supported by our analysis and data. Instead, we suggest that sXIST RNA may regulate the expression of autosomal genes involved in cilia-related processes and others that are important for neuromuscular development and in male and female peripheral nerves. lncRNA functions, however, are diverse and challenging to define (Statello et al. 2021; Mattick et al. 2023). Among many other potential mechanisms, we suggest that sXIST could act as a miRNA sponge. This hypothesis also considers the possibility of sXIST RNA existing as circular RNAs (circRNAs) because it contains short repeat sequences. In fact, many circRNAs mapped to the locus are recorded in the circAtlas (Wu et al. 2020), for example, hsa-XIST_0011. Therefore, it is possible that sXIST RNAs can sequester miRNAs that target genes in processes important for nonmyelinating Schwann cell functions, such as cell–cell signaling through primary cilia, which exist in glia and may be involved in a variety of functions related to nerve regeneration signaling (Ki et al. 2021). This is in line with the main functions of Schwann cells in supporting nerve development and regeneration after injury. We also noticed that many zinc-finger TFs were upregulated in our sXIST overexpressing cells (Supplemental Fig. S13E), but the functional implication is unclear.
Among the top 15 miRNAs predicted to target SPC genes, five of them (miR-361-3p, miR-32, miR-149-3p, miR-122, and miR-367) have been reported to interact with XIST, although in those studies the specific XIST isoforms were not addressed and were all in cancer contexts (Li et al. 2018; Cheng et al. 2020; Yang et al. 2020; Jiang et al. 2021; Wang et al. 2023).
From our sXIST overexpression data, we similarly found that three miRNAs (miR-520a-5p, miR-9, and miR-34a) predicted to target upregulated genes were previously reported to interact with XIST, with miR-520a-5p described in non-small-cell lung cancer resistance (Liu et al. 2021) and miR-34a in thyroid cancer (Liu et al. 2018). As for miR-9, it is a known regulator of neurogenesis (Coolen et al. 2013); it was shown to inhibit Schwann cell migration (Zhou et al. 2014); and it is targeted by XIST to regulate mesenchymal stem cell differentiation in the bone marrow (Zheng et al. 2020). If XIST acts as a sponge for miR-9, its downregulation in neuropathy (Supplemental Fig. S15) may exacerbate symptoms because the excessive miR-9 could lead to reduced Schwann cell migration to damaged nerves. It is therefore compelling that HMMR (also known as RHAMM or CD168) shares a binding site with miR-9-5p at its 3′ UTR (Fig. 5G). HMMR is important for the motility of Schwann/Schwann-like cells (Ouasti et al. 2020) and is involved in the PLK1-dependent pathway that stabilizes astral microtubules coming from the centrosome (Connell et al. 2017). Centrosomes and the microtubules they produce are important for primary cilia development (Dantas 2020), so we wonder if HMMR and other cilia-related genes affected by XIST expression could affect primary cilia production/maintenance.
Future studies are needed to address this and, more importantly, whether these miRNAs and others predicted from our analysis bind to sXIST RNA in Schwann cells and lead to altered expressions of their other targets, at either the transcript or protein level. We should note that miRNAs can regulate their targets both negatively and positively (Hausser and Zavolan 2014).
It is important to point out that the model of XIST functioning as a miRNA sponge will depend on its location in the cytoplasm, but some miRNAs are located in nuclei (Marshall et al. 2019; Morey et al. 2025). The full-length XIST RNA contains sequences that retain it to nuclei, but these sequences are mostly absent in the sXIST transcript. Thus, although we did not have data to resolve the subcellular location of sXIST RNA, conceivably it can be in the cytoplasm, nucleus, or both (Politz et al. 2006; Leung 2015; Turunen et al. 2019). Future studies will be needed to pinpoint sXIST’s cytoplasmic and/or nuclear distribution and determine its direct interactions with key miRNA targets.
Our finding of XIST expression changes in cardiomyopathy and PNP is also interesting. XIST and its miRNA sponging capacity are suggested in a variety of cardiovascular/circulatory system–related diseases, such as myocardial ischemia, cardiac hypertrophy, myocardial infarction, atherosclerosis, deep vein thrombosis, pulmonary arterial hypertension, and more (Almalki 2024). Gorin and Goodman (2025) similarly showed that XIST expression was higher in cardiomyopathy, but we are unaware of other literature directly linking XIST to this disease. Our results revealed that RYR2, which exhibited greater negative correlation with XIST in male peripheral nerves and therefore may be (either directly or indirectly) downregulated by the sXIST transcript, was significantly downregulated in glial cells of cardiomyopathy hearts. Because RYR2 is an important cardiac muscle–related gene involved in multiple diseases (Sleiman et al. 2021), we wonder whether ryanopathies are partly influenced by aberrant sXIST expression. If so, specifically targeting sXIST could upregulate RYR2 and modulate disease progression. However, more work is needed to prove that sXIST directly influences RYR2 expression.
Another critical question is whether the expression, regulation, and function of XIST is conserved across mammalian species. We tried to address this, but mining sex-specific gene expression from published mouse RNA-seq data is extremely difficult because almost all studies pool tissues from both sexes. We attempted to overcome this limitation by predicting the sex of individual cells, but the approach will not work for bulk RNA-seq data. Based on our analysis and some limited data in the mouse ENCODE Project, we are confident that Xist is expressed in peripheral nerves in male mice, but we cannot determine if the alternative promoter and resulting transcript(s) are conserved between humans and mice.
Finally, our analysis of the CELLxGENE data indicated that several other cell types, such as pancreatic stem cells, ciliary epithelial cells, and fibro/adipogenic progenitor cells also exhibit active XIST expression in male cells. In future studies, we need to address if sXIST is activated and functional in these cell types like in Schwann cells.
In conclusion, our study suggests that sXIST is highly specific to peripheral glia and potentially acts as a molecular sponge to sequester miRNAs that are involved in modulating various Schwann cell functions. Further work is needed to confirm the exact region of sXIST that regulates the expression of nervous system–related processes and whether this indeed occurs through miRNA binding. Although we focus our discussion on miRNA sponge function in this study, the XIST functions in peripheral glia are very likely more complex. For example, we do not know if the short and full XIST transcripts interact in female cells.
Methods
Overexpression of sXIST in the HEK-293T cell line and RNA-seq
The last 3323 bp sequence of XIST exon 6 (Chr X: 73,820,656–73,823,979(−)) was cloned into pLV-eGFP vector and transfected into HEK-293T cells for overexpression. Total RNA was isolated from the sXIST overexpression cells and controls for strand-specific RNA-seq (see Supplemental Methods). RNA-seq reads were mapped to the human genome (GRCh38) with the GENCODE annotation (v46) (Mudge et al. 2025) by the STAR aligner (v2.7.9a) (Dobin et al. 2013). Gene counts were determined using RSEM (v1.2.28) (Li and Dewey 2011). Differential expression analysis was performed using DESeq2 (Love et al. 2014).
Public bulk and single-cell data
Human peripheral nerve bulk RNA-seq data
The data were collected by Sandy-Hindmarch et al. (2025) from 11 controls (four females, seven males) and 21 Morton's neuroma samples (18 females, four males) to understand the molecular signatures of nerve injury. All Morton's neuroma samples were from the plantar digital nerves, whereas control samples were collected from a variety of upper and lower limb nerves (three plantar digital, three gracilis motor, four posterior interosseous sensory, and one intercostal sensory). We downloaded the raw count expression data and FASTQ files (generated from strand-specific RNA-seq) from the Gene Expression Omnibus (GEO), accession number GSE250152. We mapped the RNA-seq reads with STAR (Dobin et al. 2013) to the GRCh38/hg38 genome build, as the authors did. The alignment BAM files for individual samples were then merged by sex and neuroma status and subsequently indexed using SAMtools v1.14 (Danecek et al. 2021). Then, reads mapped to the XIST locus (Chr X: 73,820,656–73,852,714) were extracted and visualized in the Integrative Genomics Viewer (IGV) (Robinson et al. 2011). For expression difference between the first and sixth exon, the number of reads were obtained using SAMtools for exon 6 (Chr X: 73,820,548–73,828,166) and exon 1 (Chr X: 73,841,317–73,852,768) for each sample. The ratios of the exon 6 reads to exon 1 reads for male and female samples were compared by t-tests.
Human tibial and sciatic nerve bulk RNA-seq, ATAC-seq, and ChIP-seq data
The data were downloaded from the ENCODE Project (The ENCODE Project Consortium 2012) data portal, including RNA-seq and ChIP-seq (H3K4me3, H3K4me1, H3K27ac, H3K36me3, H3K27me3, and H3K9me3) data from the tibial nerve of 37- and 54-year-old males and ATAC-seq data from the sciatic nerve of a 26-year-old male and 16- and 47-year-old females. For positive (+)- and negative (−)-strand RNA-seq bigWig files, we used bigWigSummary from the UCSC Genome Browser (Perez et al. 2025) to get the average signal for exon 6 (Chr X: 73,820,548–73,828,166) and exon 1 (Chr X: 73,841,317–73,852,768), which we then multiplied by the exon length to get the total signal. We used these signal values to calculate the ratio of exon 6 to exon 1 signal for each strand and sample. The accession numbers for the data used for results presented in Figure 3 and Supplemental Figure S7 are listed in Supplemental Table S1.
Human heart long-read RNA-seq data
The data were downloaded from the ENCODE Project (The ENCODE Project Consortium 2012) data portal. BAM files for accession numbers ENCFF330VRX and ENCFF327SFR were downloaded and filtered for reads on the X Chromosome.
Human heart sc/snRNA-seq data
The data were from three difference sources (Supplemental Table S1; Supplemental Methods; Sim et al. 2021; Mehdiabadi et al. 2022; Kanemaru et al. 2023). These three data sets were reanalyzed using Seurat (Hao et al. 2024) (version 5) to manage sample-level metadata (age/age range, sex, and data set) and filter for cells/nuclei with more than 200 detected genes and percentage of mitochondrial gene expression < 10% for scRNA-seq data and 5% for snRNA-seq data. Next, we normalized raw counts for all sc/snRNA-seq data with Seurat's NormalizeData function, followed by the FindVariableFeatures, ScaleData, and RunPCA functions. We also used Scrublet (Wolock et al. 2019) to predict and remove doublets from these data. Next, we used reciprocal principal component analysis (RPCA) (Hao et al. 2024) to integrate data from all samples, followed by clustering for consistent cell-type identification using the original samples’ metadata and cell-type markers in the three original studies.
Human skeletal muscle atlas data
The data were obtained from https://www.muscleageingcellatlas.org/, and they were collected by Kedlian et al. (2024). We used only the “fibroblasts and Schwann cells” data set, which was converted from an h5ad object to a Seurat object in R using the anndata (https://github.com/dynverse/anndata) and sceasy (https://github.com/cellgeni/sceasy) R pacakges. These data were derived from the intercostal muscles of 12 (four females, eight males) deceased donors between the ages of 15 and 75 years. The data were already preprocessed using SCANPY (Wolf et al. 2018), in which the authors first used CellBender (Fleming et al. 2023), which removes potential technical artifacts from sc/snRNA-seq data. They then filtered for cells/nuclei with mitochondrial gene expression < 10% for scRNA-seq data and <5% for snRNA-seq data, along with other gene detection and counts filters, such as a count range between 400 and 40,000. The authors also shared their cell-type annotations, including nonmyelinating and myelinating glia and the corresponding markers (Kedlian et al. 2024).
Cardiomyopathy heart snRNA-seq data
These data were obtained from a previous study by Reichart et al. (2022). The samples were collected from donor ventricles to characterize the cell type and gene program changes that occur in hearts with arrhythmogenic right ventricular and dilated cardiomyopatheis (Reichart et al. 2022). Data for 18 controls and 61 cardiomyopathy hearts were downloaded from https://cellxgene.cziscience.com/collections/e75342a8-0f3b-4ec5-8ee1-245a23e0f7cb/private. They were uploaded by the authors and processed after filtering for counts between 300 and 15,000, 300–5000 detected genes for gene detection, and <1% mitochondrial expression. Cell-type annotation was also provided. Because there were no male cells in the noncompaction cardiomyopathy samples, we eliminated this group from our analysis, leaving us with normal, arrhythmogenic right ventricular cardiomyopathy, and dilated cardiomyopathy groups.
Mouse snRNA-seq data
These data were from multiple peripheral nerves (GEO: GSE182098; snRNA-seq) (Yim et al. 2022) and sciatic nerves (GEO: GSE137870; scRNA-seq) (Gerber et al. 2021) and available as processed Seurat objects. The data by Yim et al. (2022) came from a combination of sciatic, sural, vagus, and peroneal nerve collections. For both data sets, multiple mice, regardless of sex, were pooled. Both provided detailed assessments of glial cell types/subtypes. For Yim et al. data, we used their processed data and cell-type annotations without further filtering. For Gerber et al. data, we subclustered their labeled Schwann cells to help us differntiate myelinating from nonmyelinating cells. For the data by Gerber et al., they were preprocessed by filtering for cells with 500–5000 genes, whereas Yim et al. removed nuclei with fewer than 500 genes and with >5% mitochondrial gene expression.
Mouse and human cell atlas
The tissue and cell-type level expression data for XIST/Xist were obtained using the CELLxGENE discover gene expression tool (CZI Cell Science Program et al. 2024). We summarize all cell type–tissue combinations with XIST/Xist detection in >30% of male cells in Supplemental Figure S3 and provide the percentages for all other cell types by sex and tissues in Supplemental Tables S3 and S10. These data came from many sources and studies but were reprocessed consistently by the CELLxGENE team, including the removal of duplicated cells and cells with fewer than 500 genes and then keeping only cells derived from sequencing assays that do not require gene-length normalization, such as 10x 3′ and 5′ kits, Drop-Seq, Seq-Well, and CEL-Seq. The data can be obtained from https://cellxgene.cziscience.com/gene-expression.
Multiomic heart data
The 10x multiomic scRNA/ATAC-seq data were also obtained from the Heart Cell Atlas (Kanemaru et al. 2023). ATAC fragments and peaks from 10 (four female, six male) adult hearts (between 20 and 75 years of age) were obtained from https://www.heartcellatlas.org/. Author-provided metadata were used for labeling major cell types and sex. We used the Signac R package (Stuart et al. 2021) and followed the standard analysis procedures to merge data for each of the samples. Peaks were originally called based on high-resolution cell types that the authors annotated. We thus utilized MACS2 (Zhang et al. 2008) to recall peaks after merging data to the level of broad cell types. We also analyzed the expression data after retaining only cells present in the ATAC data set based on barcode. We then used Signac's built-in plotting functions to create ATAC coverage plots over the XIST locus.
CATlas data
The CATlas (Zhang et al. 2021) is a crowdsourcing resource that curates human and mouse cis-regulatory element reference maps. We downloaded and used bigWig files without further processing. We obtained these cell-type-level bigWig files for snRNA and ATAC-seq data from their curated heart and multitissue data sets.
sc/snRNA-seq data cell-type identification
For sc/snRNA-seq data without cell-type annotations for direct usage, we first computed cell-type markers using Seurat's FindAllMarkers function, which performs a Wilcoxon rank-sum test between cells in each cluster and all other cells. We then compared the top markers with known glia, myelination, and neuronal markers, adapted from the method of Kanemaru et al. (2023), to further identify glial subtypes (myelinating, nonmyelinating, or other) and to ensure that they were negative for neuronal markers. SCN7A was identified as a Schwann cell marker in the Human Protein Atlas (Karlsson et al. 2021) and is more highly expressed in nonmyelinating Schwann cells by Yim et al. (2022), along with SLC35F1. Thus, these genes were included as Schwann cell markers. As in Kanemaru et al. (2023), NRXN1, NRXN2, NCAM2, ALDH1A1, and S100B were used as glial cell markers. Kanemaru et al. considered MBP, MPZ, and PRX as general Schwann cell markers; however, we additionally added PLP1, which encodes a major component of myelin (Diehl et al. 1986), to specifically identify myelinating Schwann cells. We also added XIST, CADM2, and SMYD3, which were markers for cardiac nonmyelinating Schwann cells from our integrated data. NGF was used to identify “other” glia because NGF+ heart glia clustered separately. Because NRXN1 and NRXN2 are also expressed in neurons, we needed to include genes to ensure that glia were not neurons. For this, we used the same neuronal markers as Kanemaru et al., which included PRPH, STMN2, CACNA1B, NEFL, NEFM, NEFH, DLG4, and TUBB3. The markers listed here were also used to identify Schwann cells in nonheart data sets when such cell types were not defined or shared by the original authors.
Differential expression analysis
For sc/snRNA-seq data sets, pseudobulking by cell type or subcluster was performed prior to differential expression analysis with DESeq2 (Love et al. 2014) in order to use the sample replicates appropriately. For the integrated human heart data, a group variable was introduced to account for batch effects (e.g., laboratories) observed across data sets. All other parameters were set to default. For more information on density plots, gene set enrichment analysis, and overrepresentation analysis, please refer to Supplemental Methods.
Defining genes with strong expression correlation with sXIST
To predict genes that may be regulated by the sXIST transcript, we applied coexpression analysis to the human peripheral nerve bulk RNA-seq data from Sandy-Hindmarch et al. To control for the effect of expression level, we first needed to obtain a list of genes with similar levels of expression as XIST. Based on the DESeq2-normalized count data, we filtered for genes with mean expression values across samples within ±3 of XIST’s mean. After, we randomly selected 100 of such genes and calculated the Pearson's correlation between them (and XIST) and all other genes across male and female samples, separately. Thus, for each of these 101 genes, we obtained male and female correlation coefficient values with the rest of the (expressing) human genes. The values were used to prepare a female versus male correlation coefficient 2D plot. We next fit an ellipse around 99% of the data points to obtain a background null distribution. Applying this ellipse to the correlation coefficients for XIST, we obtained genes whose expression correlation with XIST were higher than the null background, namely, outliers located outside of the ellipse. We further filtered for genes with an absolute difference >0.3 between their correlation coefficients in male and female samples and grouped them into two groups: stronger negative correlation (SNC) genes (male correlation coefficient [r] < 0 and P-value < 0.05), and stronger positive correlation (SPC) genes (male correlation coefficient [r] > 0 and P-value < 0.05).
Pooled mouse scRNA-seq sex prediction
To predict each cell's sex (XX/XY) for pooled mouse sc/snRNA-seq samples, we first obtained a list of all Y Chromosome genes using the biomaRt (Durinck et al. 2009) R package. We then annotated a cell as male if it showed expression of any of the Y Chromosome genes. Changing this to a stricter criterion, for example, increasing the expression counts to three, did not affect our results. For the mixed peripheral nerve data by Yim et al., we eliminated Y Chromosome genes starting with “Gm” from this filtering step, which were abundant in the data set (perhaps owing to their abundance in snRNA-seq). To assess the positive rate of this cell-level sex prediction, we used integrated heart and the Skeletal Muscle Atlas single-cell/-nucleus data, described above.
Data access
RNA-seq data from HEK-293T cells have been submitted to the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo) under accession number GSE309419. All analysis codes in R (R Core Team 2023) are publicly available at GitHub (https://github.com/bioinfoDZ/sXIST) and as Supplemental Code.
Competing interest statement
The authors declare no competing interests.
Acknowledgments
We thank the Einstein High Performance Computing and members in the Zheng laboratory for providing valuable suggestions. This study was supported partially by the National Heart, Lung, and Blood Institute (NHLBI) grants R01HL153920 and R01HL163667 (B.E.M., D.Z.) and by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grants R01AR069078 and P30AR079200 (E.E.).
Author contributions: K.S.O. and D.Z. conceived and designed the study. K.S.O. performed all the bioinformatic data analysis, with assistance from K.J. M.-Y.L. and E.E. performed the sXIST overexpression assay. L.S. and B.E.M. contributed to the mouse data analysis. K.S.O. and D.Z. wrote the manuscript. All authors reviewed and edited the manuscript.
Footnotes
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.280832.125.
-
Freely available online through the Genome Research Open Access option.
- Received May 4, 2025.
- Accepted November 26, 2025.
This article, published in Genome Research, is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.