A low-abundance class of Dicer-dependent siRNAs produced from a variety of features in C. elegans
- Thiago L. Knittel1,4,
- Brooke E. Montgomery1,4,
- Alex J. Tate1,
- Ennis W. Deihl2,
- Anastasia S. Nawrocki2,
- Frederic J. Hoerndli2,3 and
- Taiowa A. Montgomery1,2
- 1Department of Biology, Colorado State University, Fort Collins, Colorado 80523, USA;
- 2Department of Biomedical Sciences, Colorado State University, Fort Collins, Colorado 80523, USA;
- 3Cell and Molecular Biology Program, Colorado State University, Fort Collins, Colorado 80523, USA
-
↵4 These authors contributed equally to this work.
Abstract
Canonical small interfering RNAs (siRNAs) are processed from double-stranded RNA (dsRNA) by Dicer and associate with Argonautes to direct RNA silencing. In Caenorhabditis elegans, 22G-RNAs and 26G-RNAs are often referred to as siRNAs but display distinct characteristics. For example, 22G-RNAs do not originate from dsRNA and do not depend on Dicer, whereas 26G-RNAs require Dicer but derive from an atypical RNA duplex and are produced exclusively antisense to their messenger RNA (mRNA) templates. To identify canonical siRNAs in C. elegans, we first characterized the siRNAs produced via the exogenous RNA interference (RNAi) pathway. During RNAi, dsRNA is processed into ∼23 nt duplexes with ∼2 nt, 3′-overhangs, ultimately yielding siRNAs devoid of 5′G-containing sequences that bind with high affinity to the Argonaute RDE-1, but also to the microRNA (miRNA) pathway Argonaute, ALG-1. Using these characteristics, we searched for their endogenous counterparts and identified thousands of endogenous loci representing dozens of unique elements that give rise to mostly low to moderate levels of siRNAs, called 23H-RNAs. These loci include repetitive elements, putative coding genes, pseudogenes, noncoding RNAs, and unannotated features, many of which adopt hairpin (hp) structures reminiscent of the hpRNA/RNAi pathway in flies and mice. RDE-1 competes with other Argonautes for binding to 23H-RNAs. When RDE-1 is depleted, these siRNAs are enriched in ALG-1 and ALG-2 complexes. Our results expand the known repertoire of C. elegans small RNAs and their Argonaute interactors, and demonstrate that key features of the endogenous siRNA pathway are relatively unchanged in animals.
Small interfering RNAs (siRNAs) are ∼21–24 nt long noncoding RNA produced through cleavage of double-stranded RNA (dsRNA) by the endoribonuclease Dicer (Hamilton and Baulcombe 1999; Hammond et al. 2000; Zamore et al. 2000; Bernstein et al. 2001; Elbashir et al. 2001; Grishok et al. 2001; Ketting et al. 2001; Knight and Bass 2001). Argonaute proteins associate with siRNA duplexes, discarding one strand and retaining the other, which then acts as a guide to direct silencing of target messenger RNAs (mRNAs) (Grishok et al. 2001; Hammond et al. 2001).
Shortly after the discovery that dsRNA triggers RNA silencing in Caenorhabditis elegans, siRNAs were identified in plants for their role in silencing viruses and transgenes, revealing the specificity factor of the RNA interference (RNAi) pathway (Fire et al. 1998; Hamilton and Baulcombe 1999). In plants, beyond their roles in silencing viruses and transgenes, siRNAs also target endogenous coding genes, intergenic regions, and transposons (Llave et al. 2002; Kasschau et al. 2007). Similarly, in mice and flies, endogenous siRNAs originate from various sources, most notably transposons, suggesting a common role for siRNAs in silencing foreign elements (Chung et al. 2008; Czech et al. 2008; Ghildiyal et al. 2008; Okamura et al. 2008a,b; Tam et al. 2008; Watanabe et al. 2008).
C. elegans contains multiple distinct classes of endogenous small RNAs but canonical siRNAs like those found in other species have not been extensively studied. What is often classified as siRNAs in C. elegans lacks key features characteristic of siRNAs in other species (Claycomb 2014). For example, 22G-RNAs do not require Dicer and appear to lack a dsRNA intermediate (Aoki et al. 2007; Pak and Fire 2007; Gu et al. 2009; Claycomb 2014). 26G-RNAs are similar to canonical siRNAs in their requirement for Dicer, but these small RNAs derive from an asymmetric dsRNA intermediate structure that lacks the ∼2 nt, 3′-overhangs characteristic of Dicer products (Ambros et al. 2003; Ruby et al. 2006; Han et al. 2009; Conine et al. 2010; Gent et al. 2010; Vasale et al. 2010; Welker et al. 2010; Fischer et al. 2011; Blumenfeld and Jose 2016). Both 22G-RNAs and 26G-RNAs are produced antisense to their mRNA templates through the activity of RNA-dependent RNA polymerases, which further distinguishes them from siRNAs in mice and flies, but is a common feature of most siRNAs in plants and fungi (Cogoni and Macino 1999; Dalmay et al. 2000; Mourrain et al. 2000; Smardon et al. 2000; Sijen et al. 2001; Volpe et al. 2002; Aoki et al. 2007; Pak and Fire 2007; Gent et al. 2009, 2010; Gu et al. 2009; Han et al. 2009; Conine et al. 2010; Vasale et al. 2010).
Both exogenous RNAi and viral infection trigger a silencing response involving the production of exogenous canonical siRNAs in C. elegans (Ketting et al. 2001; Ashe et al. 2013). Furthermore, studies exploring the interplay between RNAi pathways and RNA editing by adenosine deaminases (ADARs) in C. elegans have identified numerous endogenous small RNAs with the hallmarks of canonical siRNA. For example, they appear to be processed from a dsRNA intermediate and are 22–24 nt long (Wu et al. 2011; Reich et al. 2018; Fischer and Ruvkun 2020). These siRNAs are highly upregulated in animals with loss of function in one or both ADAR genes, presumably because in wild-type animals the base changes introduced by RNA editing disrupt the secondary structure of dsRNA, preventing its recognition or processing by Dicer (Wu et al. 2011; Reich et al. 2018; Fischer and Ruvkun 2020).
Endogenous siRNAs may constitute a distinct class of small RNAs largely overlooked in C. elegans. Presumably, such siRNAs are rare in wild-type C. elegans since they have not emerged from high-throughput small RNA sequencing (sRNA-seq) studies characterizing small RNAs (Ruby et al. 2006). Nonetheless, even at low abundance, they could have important roles in gene regulation and thus their identification would be an important step toward a more complete understanding of the small RNA landscape of C. elegans. In this study, our aim was to identity and characterize these siRNAs in the context of their Argonaute binding partners.
Results
Molecular attributes of exogenous siRNAs in C. elegans
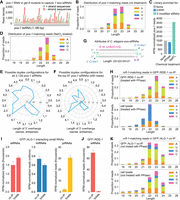
In C. elegans, siRNAs are processed from exogenous dsRNA but accumulate at very low levels compared to the secondary 22G-RNAs, which are produced downstream by RNA-dependent RNA polymerases (Sijen et al. 2001; Pak and Fire 2007). Because 22G-RNAs are produced antisense to an mRNA template, sRNA-seq can be used to distinguish primary siRNAs from secondary 22G-RNAs by their alignment in the sense orientation to target mRNAs (Sijen et al. 2001; Pak and Fire 2007). Although antisense reads are necessarily lost with this approach, we and others have used it to characterize exogenous siRNAs as predominantly 21–24 nt with a preference for a 5′U, A, or C (Zhang et al. 2012; Svendsen et al. 2019). However, the presence of 22 nt, 5′G-containing reads aligning to the sense strand of the mRNA in these libraries suggests contamination from 22G-RNAs, complicating the distinction between primary siRNAs and 22G-RNAs. To further refine the molecular attributes of exogenous siRNAs, we analyzed sRNA-seq libraries from glp-4 mutants treated with RNAi against the germline-expressed gene, pos-1 (Svendsen et al. 2019). Because glp-4 mutants lack a germline when grown at 25°C, they express very little pos-1 mRNA and thus have minimal capacity for 22G-RNA production (Beanan and Strome 1992). Therefore, pos-1 aligning sequences will correspond almost exclusively to the primary siRNAs produced from the exogenous dsRNA (Fig. 1A). Consistent with our previous findings, primary siRNAs were predominantly 22–23 nt long and depleted of 5′G-containing sequences (Fig. 1B; Zhang et al. 2012; Svendsen et al. 2019).
Attributes of exogenous siRNAs in C. elegans. (A) siRNA sequences matching exogenously delivered pos-1 dsRNA in glp-4(bn2) mutants as identified using sRNA-seq. n = 1 biological replicate. Libraries from adults treated with pos-1 RNAi and grown at 25°C to induce sterility. (B) Size and 5′ nt distribution of pos-1-matching reads from the sRNA-seq libraries in A. (C) RPM-normalized pos-1-matching reads from untreated (none), RNA 5′ polyphosphatase-treated (PPase, reduces 5′ triphosphates), and NaIO4-treated (blocks ligation of non-3′-end-modified small RNAs) sRNA-seq libraries. n = 1 biological replicate. RNA is the same as in the library in (A,B). (D) Size and 5′ nt distribution of pos-1-matching reads from the NaIO4-treated sRNA-seq library in C. (E) The numbers of pos-1-matching siRNA sequences with the possible duplex configurations indicated. Note that a single sequence could have multiple possible complementary strands with different 3′-overhangs. (F) As in E but limited to siRNAs with ≥50 reads. (G) Attributes of exogenous siRNAs in C. elegans. (H) Size and 5′-nt distribution of nrfl-1-matching reads from GFP::RDE-1 co-IP and cell lysate sRNA-seq libraries. n = 2 biological replicates. Data from one representative library for each condition are shown. Libraries from gravid adults treated with nrfl-1 and oma-1 RNAi. (I) RPM-normalized miRNA, piRNA, and nrfl-1-matching reads in GFP::ALG-1 co-IP and cell lysate sRNA-seq libraries. Libraries from gravid adults treated with nrfl-1 RNAi. Error bars are standard deviation (SD) between three biological replicates. (J) RPM-normalized nrfl-1-matching reads in GFP::RDE-1 co-IP and cell lysate sRNA-seq libraries. Libraries from gravid adults treated with nrfl-1 RNAi. Error bars are SD between three biological replicates. (K) Size and 5′-nt distribution of nrfl-1-matching reads from GFP::ALG-1 co-IP and cell lysate sRNA-seq libraries. Data from one representative library for each condition are shown.
22G-RNAs contain triphosphates at their 5′ ends, whereas primary siRNAs contain monophosphates (Pak and Fire 2007). Enzymatic treatment of small RNAs to reduce triphosphates to monophosphates before library construction can be used to enrich for 22G-RNAs, and conversely, excluding this treatment can be used to enrich for primary siRNAs (Almeida et al. 2019). Pretreatment of small RNA libraries from pos-1-treated glp-4-mutants with RNA 5′ polyphosphatase to reduce triphosphates to monophosphates had little impact on the relative abundance of pos-1-matching reads (Fig. 1C). This is further evidence that these libraries have very low levels of 22G-RNAs produced from pos-1 and confirms that exogenous siRNAs are monophosphorylated.
We previously showed that exogenous siRNAs are modified at their 3′ ends by the methyltransferase HENN-1 (Svendsen et al. 2019). We observed an approximately twofold enrichment of pos-1-matching reads in libraries pretreated with sodium periodate (NaIO4), an oxidizing agent that selectively targets unmodified 3′ bases, inhibiting 3′ adapter ligation to these sequences (Fig. 1C; Seitz et al. 2008; Yu and Chen 2010; Svendsen et al. 2019). NaIO4 treatment further enriched for 23 nt sequences and depleted 5′G-containing sequences, likely due to the loss of residual 22G-RNAs present in glp-4 mutants (Fig. 1D).
Canonical siRNAs are processed from longer dsRNA into ∼21–24 nt duplexes by the RNaseIII-like enzyme Dicer (Bernstein et al. 2001; Elbashir et al. 2001; Ketting et al. 2001). Like other RNaseIII cleavage products, siRNA duplexes typically have 2 nt, 3′-overhangs (Robertson et al. 1968; Elbashir et al. 2001). Although viral siRNA duplexes commonly have 2 nt overhangs in C. elegans, in cell-free extracts, dsRNA is often processed into duplexes with 3–4 nt overhangs (Welker et al. 2011; Ashe et al. 2013). To determine the most prevalent duplex configuration in exogenous RNAi, we identified all possible pos-1 siRNA duplexes produced during RNAi. To minimize contamination of 22G-RNAs, we examined sRNA-seq data from libraries treated with NaIO4, since these libraries had the lowest levels of 22 nt, 5′G-containing reads (Fig. 1D). Even for the most abundant potential duplexes, one strand was always present at low levels, suggesting that strand stabilization is very selective (Supplemental Fig. S1A). Among the possible 0–4 nt, 3′-end overhangs, configurations with 2 nt overhangs on either strand of the duplex were most prevalent, particularly when only including possible duplexes in which one strand had ≥50 reads (Fig. 1E,F).
Since complementary sequences may not originate from the same duplex, we cannot confirm Dicer's preference for siRNA duplexes with specific 3′ overhangs in vivo. Moreover, stabilization of certain duplexes or single strands postdicing may skew results. However, our data suggest Dicer processes dsRNA imprecisely, producing various 3′ overhangs, with 2 nt overhangs being the most common. In summary, we conclude that exogenous siRNAs have the following attributes: a predominant length of 23 nt, a bias against 5′G, a 5′ monophosphate, a duplex intermediate with ∼2 nt, 3′ overhangs, and a 2′-O-methyl group at their 3′ ends (Fig. 1G).
RDE-1 and ALG-1 associate with exogenous siRNAs
RDE-1 associates with at least some exogenous siRNAs and is essential for RNAi (Tabara et al. 1999; Yigit et al. 2006). To more comprehensively assess the extent to which RDE-1 binds exogenous siRNAs, we co-immunoprecipitated (co-IP'd) GFP::RDE-1-small RNA complexes from animals undergoing RNAi against both nrfl-1 and oma-1, genes chosen for their mild phenotypes (Davis et al. 2022). Animals in this experiment, unlike in the pos-1 RNAi experiments above, were able to mount a 22G-RNA amplification response since the endogenous targets were present. Small RNAs matching nrfl-1 or oma-1 in the cell lysate input fractions used for GFP::RDE-1 co-IPs were predominantly 22 nt and strongly biased for 5′G and were thus comprised mainly of 22G-RNAs (Fig. 1H; Supplemental Fig. S1B). In contrast, nrfl-1- and oma-1-matching reads from the GFP::RDE-1 co-IP fractions were predominantly 23 nt and were depleted of reads containing 5′G (Fig. 1H; Supplemental Fig. S1B).
siRNA and microRNA (miRNA) duplexes are structurally similar except that miRNA duplexes typically have internal mispairs. While some miRNAs bind RDE-1, they primarily function with ALG-1 and ALG-2, and to a lesser extent with ALG-5 (Grishok et al. 2001; Corrêa et al. 2010; Brown et al. 2017; Seroussi et al. 2023). Since RDE-1 interacts with both siRNAs and miRNAs, this raises the possibility that other miRNA-associated Argonautes, such as ALG-1, may also bind siRNAs. To assess this, we sequenced small RNAs that co-IP'd with GFP::ALG-1 in animals undergoing nrfl-1 RNAi. While nrfl-1-matching siRNAs were less enriched than miRNAs in GFP::ALG-1 co-IPs, based on reads per million (RPM) normalization relative to the cell lysates, they were substantially more enriched than PIWI-interacting RNAs (piRNAs), which associate with PRG-1 and therefore represent carryover from the cell lysate (Fig. 1I; Batista et al. 2008; Das et al. 2008; Wang and Reinke 2008; Seroussi et al. 2023).
Despite their association with GFP::ALG-1, the relative abundance and enrichment of nrfl-1-matching siRNAs were significantly lower compared to a parallel set of GFP::RDE-1 co-IPs, likely reflecting ALG-1's broader association with miRNAs (Fig. 1J; Brown et al. 2017; Seroussi et al. 2023). nrfl-1 siRNAs were predominantly 23 nt in cell lysates used for GFP::RDE-1 and GFP::ALG-1 co-IPs (Fig. 1K; Supplemental Fig. S1C). These samples were not treated with RNA 5′ polyphosphatase, hence the absence of 22G-RNAs in these libraries. siRNA reads were shifted slightly toward 22 nt sequences in GFP::ALG-1 co-IPs, but further toward 23–24 nt sequences in GFP::RDE-1 co-IPs, suggesting that the 23 nt bias may be driven by RDE-1 preferentially binding longer small RNAs (Fig. 1K; Supplemental Fig. S1C). Both Argonaute co-IPs displayed a lack of 5′G-containing siRNAs, implying either a shared bias or other factors influencing the 5′ nt (Fig. 1K; Supplemental Fig. S1C). These results show that both RDE-1 and ALG-1 associate with exogenous siRNAs. However, since RDE-1 is specifically required for RNAi, the role of ALG-1 in this pathway, if any, is unclear (Tabara et al. 1999).
Identification of endogenous canonical siRNAs
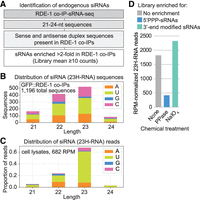
Using the features of exogenous siRNAs defined above, we set out to identify their endogenous counterparts. We analyzed GFP::RDE-1 co-IP sRNA-seq libraries, expecting them to be enriched for endogenous siRNAs due to RDE-1's strong affinity for exogenous siRNAs. We filtered these libraries for 21–24 nt reads, a size range that would capture most exogenous siRNAs. A custom algorithm was used to identify complementary sequence pairs with 2 nt, 3′ overhangs, which we predicted based on our analysis of exogenous siRNAs would most effectively capture endogenous siRNA duplexes. We further filtered for sequences with at least 10 reads and showing greater than twofold enrichment in GFP::RDE-1 co-IPs compared to cell lysates (Fig. 2A).
Identification of endogenous canonical siRNAs. (A) Canonical siRNA discovery flowchart. (B) Size and 5′ nt distribution of all endogenous canonical siRNA (23H-RNA) sequences from GFP::RDE-1 co-IP sRNA-seq libraries. n = 2 biological replicates. Data from one representative gravid adult library are shown. (C) Size and 5′ nt distribution of 23H-RNA reads in the cell lysates used for the GFP::RDE-1 co-IP in B. (D) 23H-RNA reads from untreated (none), RNA 5′ polyphosphatase-treated (PPase, reduces 5′ triphosphates), and NaIO4-treated (blocks ligation of non-3′-modified small RNAs) sRNA-seq libraries. n = 1 biological replicate. Libraries from wild-type gravid adults grown at 25°C.
To broaden our search, we included additional RDE-1-associated sequences that aligned with loci producing the small RNAs identified above, without requiring that we identify a 2 nt, 3′-overhang duplex intermediate. This approach yielded 1196 candidate sequences perfectly matching 25,701 genomic loci (reflecting the repetitive nature of many of them, as described below) (Supplemental Table S1). Similar to exogenous siRNAs, these small RNAs were depleted of 5′G-containing sequences and enriched for 23 nt sequences (Fig. 2B). Cell lysates also showed this 23 nt enrichment and 5′G depletion in reads aligning to candidate regions (Fig. 2C). Given their length and bias against 5′G-containing sequences, we named these endogenous siRNAs 23H-RNAs, where H represents U, A, or C, following the nomenclature of other small RNA classes in C. elegans (Ruby et al. 2006; Gu et al. 2009; Han et al. 2009; Reich et al. 2018).
RNA 5′ polyphosphatase treatment did not enrich for endogenous 23H-RNAs in wild-type sRNA-seq libraries, indicating that they do not contain a triphosphate group and are, therefore, likely monophosphorylated (Fig. 2D; Svendsen et al. 2019). In contrast, 23H-RNAs were moderately enriched in NaIO4-treated libraries, indicating that their 3′ ends are blocked, presumably by 2′-O-methylation (Fig. 2D; Svendsen et al. 2019). These results are consistent with the attributes we observed for exogenous siRNAs above.
23H-RNAs are produced from a variety of features, including hpRNAs and repetitive elements
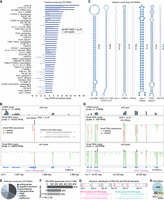
Three genomic loci—F43E2.6, Y57G11C.1145, and mir-5592—accounted for ∼98% of 23H-RNA reads in our GFP::RDE-1 co-IP sRNA-seq libraries (Fig. 3A; Supplemental Table S1). F43E2.6, which is annotated as a coding gene but with no known function, produces siRNAs from both strands, indicating that there is overlapping transcription at the locus. However, insufficient high-throughput mRNA sequencing (mRNA-seq) reads prevented a detailed analysis (Fig. 3B; Warf et al. 2012; Reed et al. 2020). Y57G11C.1145 is a long hairpin RNA (hpRNA), reminiscent of hpRNAs found in flies and mice (Fig. 3A,C; Czech et al. 2008; Kawamura et al. 2008; Okamura et al. 2008b; Tam et al. 2008; Watanabe et al. 2008). Several other 23H-RNA loci form hairpin structures that range in length from tens to hundreds of base pairs (Fig. 3C).
Genomic origins of 23H-RNAs. (A) Mean log2-RPM-normalized 23H-RNA reads from GFP::RDE-1 co-IP and cell lysate sRNA-seq libraries. Only features producing 23H-RNAs with >10 RPM in GFP::RDE-1 co-IPs are shown. Error bars are SD between two biological replicates. Libraries are the same as in Figure 2A. (B) mRNA-seq read density from wild-type animals and sRNA-seq read density from GFP::RDE-1 co-IP and cell lysate libraries plotted along the F43E2.6 23H-RNA locus. Brackets span 23H-RNA clusters. The lower track shows individual sequences but is truncated due to space constraints. The sequence of the most abundant duplex is shown. Purple bars span DCR-1-binding sites based on DCR-1 PAR-CLIP data from Rybak-Wolf et al. (2014). (C) Secondary structures for several 23H-RNA loci that form hairpins. (D) As in B but for the gene Y17D7C.3. (E) The proportion of 23H-RNA loci with >10 RPM in GFP::RDE-1 co-IPs overlapping the indicated features. (F) 23H-RNA sequences are categorized by the number of perfectly matching genomic loci. (G) PALTA3 and PAL5A genomic distribution and the most abundant duplex from each repetitive element. (H) Overlap between 23H-RNA loci and EERs and ARLs. Numbers shown are for total features; 449 23H-RNAs and EERs overlap; 129 23H-RNAs and ARLs overlap; 93 EERs and ARLs overlap; and 24 in common to all three data sets.
mir-5592 is classified as a miRNA but is unusual in having a perfectly base-paired stem and in generating multiple distinct small RNAs (Fig. 3C; Supplemental Table S1). Most 23H-RNA candidates from the locus, including miR-5592, were depleted in animals with partial loss of function in PASH-1, a protein required for processing primary miRNA transcripts (Supplemental Fig. S2A; Supplemental Table S2; Montgomery et al. 2024). Similarly, most 23H-RNAs produced from the Y57G11C.1145 hairpin locus were also reduced in pash-1 mutants (Supplemental Fig. S2A; Supplemental Table S2). However, 23H-RNAs produced from other hairpins, such as PALTA3 repetitive elements and the noncoding RNA rncs-1, were not depleted in pash-1 mutants, nor were 23H-RNAs from non-hairpin-forming loci (Supplemental Fig. S2A; Supplemental Table S2; Hellwig and Bass 2008). It is possible that mir-5592 and Y57G11C.1145 are incipient miRNAs, recognized by the processing machinery but not fully refined. mir-5550-derived small RNAs were also retained in our 23H-RNA list as they originate from a repetitive element, which is atypical of miRNAs but common among 23H-RNAs, as described below (Supplemental Table S1). We identified reads from perfectly base-paired duplexes at several other miRNA loci but largely excluded them due to uncertainty in their classification.
23H-RNAs were often produced from among clusters of other small RNAs, but only the 23H-RNAs were enriched in GFP::RDE-1 co-IPs (Fig. 3B,D; Supplemental Fig. S2B). Putative protein-coding genes, such as F43E2.6, represented ∼33% of 23H-RNA loci yielding >10 RPM in GFP::RDE-1 co-IPs, although it is unclear if they encode functional proteins (Fig. 3E). Another ∼19% of these relatively high 23H-RNA-yielding loci are annotated as pseudogenes (Fig. 3E). The second largest source of 23H-RNAs was repetitive elements with ∼28% produced from loci with >2 genomic copies and ∼4% from loci with >100 copies (Fig. 3E–G). These loci are largely related to DNA transposons, such as CELE14B, DNA-8-1, PAL8A, PALTA3, PALTA4, PAL5A, and TC5A, pointing to a possible role for 23H-RNAs in transposon silencing (Fig. 3A,G).
Dicer is required for 23H-RNA biogenesis
Most loci producing relatively high levels of 23H-RNAs were identified as Dicer substrates in PAR-CLIP experiments, indicating that 23H-RNAs are processed through Dicer cleavage as anticipated based on their duplex structures (Fig. 3B,D; Supplemental Table S3; Rybak-Wolf et al. 2014). Additionally, total RPM-normalized levels of 23H-RNAs decreased by ∼40% following dcr-1 RNAi compared to control treatment (Supplemental Fig. S2C). Although most individual 23H-RNAs were modestly depleted following dcr-1 RNAi, only a small subset passed our P-value threshold of 0.05, likely due to their low representation in our libraries (Supplemental Fig. S2D; Supplemental Table S4). We observed a similarly modest reduction in miRNA levels in these libraries (Supplemental Fig. S2E; Supplemental Table S4). The comparable effect of dcr-1 RNAi on miRNAs, which are known Dicer substrates, and 23H-RNAs further supports a role for Dicer in 23H-RNA biogenesis (Grishok et al. 2001; Hutvágner et al. 2001; Ketting et al. 2001).
Dicer's helicase domain is required for processing certain endogenous small RNAs, particularly 26G-RNAs (Welker et al. 2010). The helicase domain is important for processing dsRNA containing blunt or single-stranded 5′-end overhangs, like 26G-RNAs, but not for dsRNA with single-stranded 3′-end overhangs, such as miRNAs (Welker et al. 2011). To determine if 23H-RNAs are dependent on Dicer's helicase domain, we analyzed sRNA-seq data from dcr-1 mutants rescued with either wild-type dcr-1 or a helicase mutant form of the gene (Welker et al. 2010, 2011). 23H-RNAs were only modestly depleted in animals containing the dcr-1 helicase mutant, similar to some non-dcr-1-dependent small RNAs, in contrast to ERGO-1 class 26G-RNAs, which were almost completely lost (Supplemental Fig. S2F). Most 23H-RNA loci, including long dsRNA hairpins, produced only a few dominant duplexes, often only one or two (Fig. 3B,D,G; Supplemental Fig. 2B). Since the helicase domain is thought to promote Dicer processivity and 23H-RNAs are not strongly dependent on this domain, their biogenesis is likely not processive (Welker et al. 2011). This may explain why so few distinct 23H-RNAs are produced from each locus.
23H-RNA expression varies by developmental stage
We next compared the abundance of 23H-RNAs in gravid adults, the stage used for their identification, with L4 stage larvae, dissected distal gonads, and embryos. Most 23H-RNAs were significantly more abundant in embryos compared to adults (Supplemental Fig. S3A; Supplemental Table S5). dsRNA regions enriched in RNA editing also tend to be more highly expressed in embryos, which may reflect greater expression of transcripts forming dsRNA in embryos than in other developmental stages (Reich et al. 2018). Many 23H-RNAs also had elevated levels in the L4 stage relative to adults, and a small subset was more abundant in dissected gonads compared to whole adult animals (Supplemental Fig. S3B,C; Supplemental Table S5).
Overlap between 23H-RNAs and RNA-edited loci
We then compared 23H-RNA loci with RNA editing-enriched regions (EERs) from Reich et al. (2018) and ADAR-modulated RNA loci (ARLs) from Wu et al. (2011), which also produce siRNAs similar to 23H-RNAs but that are largely restricted to ADAR mutants (Wu et al. 2011; Reich et al. 2018). ADARs act posttranscriptionally, converting adenosine to inosine in dsRNA (Walkley and Li 2017). While extensive RNA editing typically prevents Dicer processing, limited editing can, in some instances, still permit siRNA production, albeit at low levels (Zamore et al. 2000; Scadden and Smith 2001; Warf et al. 2012). Clustering 23H-RNA loci that were within 1000 bp of each other resulted in 2998 distinct regions, of which, 449 overlapped with the 1523 EERs and 129 overlapped with the 454 ARLs (Fig. 3H; Supplemental Table S6); 2444 (∼82%) 23H-RNA loci did not overlap with these previously reported siRNA clusters. Our focus on RNA editing-proficient animals likely explains the lack of overlap of 23H-RNAs with EER and ARL siRNA clusters, while the prior studies missed siRNA clusters from less edited sites, which presumably accounts for many of the loci identified here (Wu et al. 2011; Reich et al. 2018).
To check for RNA editing in our data, we realigned sRNA-seq reads from pos-1 RNAi libraries and GFP::RDE-1 co-IP libraries allowing up to three genome mispairs. The two most abundant pos-1 siRNAs had only ∼3% and 0% of reads with A-to-G mismatches, which are indicative of RNA editing (Supplemental Fig. S4A). Approximately 22% of 23H-RNA reads from GFP::RDE-1 co-IP and cell lysate libraries had a single A-to-G mismatch and <2% had 2–3 mismatches (Supplemental Fig. S4B). miRNAs had only ∼2.5% with 1 mismatch and <0.03% with 2–3 mismatches (Supplemental Fig. S4B). Exogenous nrfl-1 siRNAs had similarly low levels, with only ∼3% having 1 mismatch and ∼0.1% having 2–3 mismatches (Supplemental Fig. S4B). sRNA-seq reads from most individual 23H-RNAs and miRNAs had <5% A-to-G mismatches, although some showed much higher levels (Supplemental Fig. S4C; Supplemental Table S7). However, many with seemingly high proportions of A-to-G mismatches were likely due to misalignment of more abundant reads from an alternative small RNA locus (Supplemental Fig. S4C). Nonetheless, for F43E2.6, for example, which was previously shown to produce siRNAs from an edited precursor, ∼39% sense-strand 23H-RNA reads had 1–3 A-to-G mismatches, which were mostly indicative of a single edit and could be unambiguously assigned to the locus (Supplemental Fig. S4D; Warf et al. 2012). In contrast, only ∼8%–9% of F43E2.6 antisense 23H-RNA reads had A-to-G mismatches (Supplemental Fig. S4D). The difference between these two strands of the same duplex may relate to the number of editable sites. Non-A-to-G mismatches accounted for 5%–7% of both F43E2.6 sense and antisense strand reads, which were likely due to 3′-end tailing and library preparation mutations (Supplemental Fig. S4D). These results indicate that endogenous 23H-RNAs can occasionally arise from lightly edited dsRNA (mostly a single A-to-G edit), whereas exogenous siRNAs are generally not produced from edited dsRNA.
RDE-1 has a high affinity for 23H-RNAs
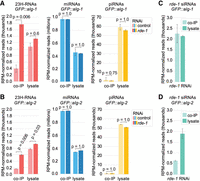
Although RDE-1 primarily associates with miRNAs, ∼14% of the total sRNA-seq reads from GFP::RDE-1 co-IPs corresponded to the 23H-RNA sequences annotated here, despite these small RNAs making up a tiny fraction of reads in the cell lysates (Fig. 4A). Although 23H-RNAs were a small percentage of total RDE-1 associated small RNAs, individual 23H-RNAs were strongly enriched in GFP::RDE-1 co-IPs compared to most miRNAs, with enrichment levels comparable to nrfl-1 and oma-1 exogenous siRNAs (Fig. 4B; Supplemental Table S8). This suggests that RDE-1 has a higher affinity for canonical siRNAs than for most miRNAs and other small RNAs.
23H-RNAs have a high affinity for RDE-1. (A) Percentages of RPM-normalized reads in GFP::RDE-1 co-IP and cell lysate sRNA-seq libraries corresponding to each class of small RNAs. n = 2 biological replicates. Libraries are the same as in Figure 2A. (B) Scatter plots display individual small RNA features as the average log2-geometric mean-normalized reads in cell lysates (x-axis) and GFP::RDE-1 co-IPs (y-axis) colored by small RNA class. Exogenous siRNAs matching nrfl-1 and oma-1 are circled. (C) RPM-normalized 23H-RNA reads in wild-type and rde-1(ne219) mutant sRNA-seq libraries. Libraries from gravid adults. Error bars are SD between three biological replicates. The P-value was calculated using a two-sample t-test. (D) Relative levels of the most abundant F43E2.6 siRNA in wild-type and rde-1Δ(ram40) mutants as determined by qRT-PCR and normalized to miR-1 levels. RNA from gravid adults. Error bars are SD between three biological replicates. The P-value was calculated using a two-sample t-test.
Despite their strong association with RDE-1, 23H-RNAs were not depleted in rde-1(ne219) mutants but instead appeared somewhat elevated (Fig. 4C). The rde-1(ne219) allele is a point mutation and could possibly still produce a protein with the ability to bind and stabilize 23H-RNAs (Tabara et al. 1999). To confirm that 23H-RNAs are not dependent on rde-1 for their stability, we generated a complete deletion allele of rde-1 (ram40). Quantitative real-time PCR (qRT-PCR) analyses showed a modest increase in the level of an F43E2.6 siRNA in the deletion allele, confirming that rde-1 is not required for the stability of 23H-RNAs (Fig. 4D).
In the absence of rde-1, 23H-RNAs might be stabilized through interactions with other Argonautes, such as ALG-1, which we showed also binds exogenous siRNAs. To identify other Argonautes that might bind and stabilize 23H-RNAs, we examined co-IP sRNA-seq data from Seroussi et al. (2023) for Argonautes involved in Dicer-dependent small RNA pathways (RDE-1, ALG-1, ALG-2, ALG-4, ALG-5, and ERGO-1) (Seroussi et al. 2023). Using DESeq2 for normalization (which involves calculating the ratio of each small RNA's counts to the geometric mean of counts for that small RNA across all samples and then normalizing by the median of these ratios), most 23H-RNAs were enriched in GFP::RDE-1 co-IPs in these data sets, as well as in co-IPs from the major miRNA Argonautes, ALG-1 and ALG-2 (Supplemental Fig. S5A–C; Supplemental Table S9; Love et al. 2014). A small subset of 23H-RNAs was also enriched in the germline-specific miRNA Argonaute, ALG-5 (Supplemental Fig. S5D; Supplemental Table S9; Seroussi et al. 2023). Some were also enriched in ERGO-1, but not in ALG-4, both of which bind 26G-RNAs, which could relate to the overlap between ERGO-1 class 26G-RNA and 23H-RNA loci, as described below (Supplemental Fig. S5E,F; Supplemental Table S9).
When measured as a proportion of RPM-normalized reads, 23H-RNAs were only collectively enriched in GFP::RDE-1 co-IPs, relative to cell lysates, although this normalization method does not account for the distribution of individual 23H-RNAs across samples (Supplemental Fig. S5G). This suggests that while other Argonautes can bind 23H-RNAs, RDE-1 has a much higher affinity for this class of small RNAs. It is also important to note that the vastly different small RNA profiles of cell lysates and Argonaute co-IPs may introduce normalization artifacts, potentially confounding these results. Nonetheless, 23H-RNAs bind preferentially to RDE-1 but also broadly interact with ALG-1 and ALG-2, which we confirm below.
Competition between Argonautes for 23H-RNAs
We hypothesized that RDE-1 competes with other Argonautes for 23H-RNAs and tested this by assessing whether the binding affinities of ALG-1, ALG-2, ALG-5, and ERGO-1, which each showed enrichment for some 23H-RNAs, were elevated in the absence of RDE-1. We treated animals with rde-1 RNAi and then co-IP'd each of the other Argonautes. By western blot, we detected an ∼90% reduction in RDE-1 protein levels following rde-1 RNAi, indicating that rde-1, despite being required for RNAi, was effectively knocked down (Supplemental Fig. S6A). In GFP::ALG-1 and GFP::ALG-2 co-IPs from animals treated with control RNAi, F43E2.6 23H-RNAs were enriched ∼16-fold and approximately eightfold, respectively, as measured by qRT-PCR, confirming that both ALG-1 and ALG-2 bind 23H-RNAs (Supplemental Fig. S6B). Following rde-1 RNAi, F43E2.6 23H-RNAs were further enriched to ∼256-fold in GFP::ALG-1 co-IPs and ∼100-fold in GFP::ALG-2 co-IPs, relative to cell lysates (Supplemental Fig. S6B). F43E2.6 23H-RNAs were not substantially enriched in GFP::ALG-5 or GFP::ERGO-1 co-IPs regardless of RNAi-treatment, indicating that these Argonautes do not compete with RDE-1 for F43E2.6 23H-RNAs (Supplemental Fig. S6B).
To determine if ALG-1 and ALG-2 more generally compete with RDE-1 for 23H-RNAs, we performed sRNA-seq on co-IPs and cell lysates from control and rde-1 RNAi-treated animals. While most small RNA reads (∼97%–98%) from GFP::ALG-1 and GFP::ALG-2 co-IPs were miRNAs, a small fraction corresponded to 23H-RNAs (Fig. 5A,B). In animals treated with rde-1 RNAi, 23H-RNA reads from GFP::ALG-1 and GFP::ALG-2 co-IPs were elevated ∼4.5- and ∼3.6-fold, respectively, relative to animals treated with control RNAi, based on RPM-normalization (Fig. 5A,B). In contrast, miRNA levels in GFP::ALG-1 and GFP::ALG-2 co-IPs were unchanged between rde-1 and control RNAi, suggesting that there is competition specifically for 23H-RNAs and not miRNAs (Fig. 5A,B). Likewise, piRNA levels were unchanged, and given that they bind PRG-1, their presence in the co-IPs likely reflects the level of small RNA carryover from cell lysates rather than an authentic interaction (Fig. 5A,B). Although 23H-RNAs were present in the co-IPs far above what would be expected due to carryover from the cell lysates based on what we observed for piRNAs, they were not collectively enriched relative to the cell lysates, except in GFP::ALG-1 co-IPs from rde-1 RNAi-treated animals (Fig. 5A,B). This likely relates to the high affinity of ALG-1 and ALG-2 for miRNAs, which were enriched ∼2.5-fold in co-IPs relative to cell lysates (Fig. 5A,B).
Competition between Argonautes for 23H-RNAs. (A,B) RPM-normalized reads corresponding to 23H-RNAs, miRNAs, and piRNAs from GFP::ALG-1 (A) or GFP::ALG-2 (B) co-IP and corresponding cell lysate sRNA-seq libraries. Two-sample t-tests were used to calculate the P-values. Bonferroni corrections were applied to account for multiple comparisons. Libraries from gravid adults treated with control (L4440) or rde-1 RNAi. Error bars are SD between two biological replicates. (C,D) RPM-normalized rde-1-matching reads from GFP::ALG-1 (C) or GFP::ALG-2 (D) co-IP and corresponding cell lysate sRNA-seq libraries.
Most individual 23H-RNAs were elevated in both GFP::ALG-1 and GFP::ALG-2 co-IPs relative to cell lysates based on geometric mean normalization, consistent with our earlier analysis (Supplemental Fig. S6C,D; Supplemental Table S10; Love et al. 2014). Many individual 23H-RNAs were enriched in co-IPs from rde-1 RNAi-treated animals relative to control treatment but were largely unchanged in cell lysates (Supplemental Fig. S6C,D). These results suggest that ALG-1 and ALG-2 compete with RDE-1 for 23H-RNAs, although ALG-1 and ALG-2 seem to have similar or lower affinities for 23H-RNAs compared to miRNAs which may limit their interactions (Supplemental Fig. 6C,D). rde-1-matching siRNAs co-IP'd with GFP::ALG-1 and GFP::ALG-2 in the animals treated with rde-1 RNAi, supporting our earlier findings that ALG-1 binds exogenous siRNAs and revealing that ALG-2 also binds siRNAs (Fig. 5C,D; Supplemental Fig. 6C,D).
Multiple small RNA pathways converge on 23H-RNA loci
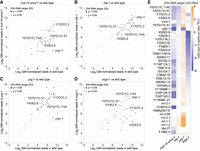
During exogenous RNAi, RDE-1 directs target mRNAs into a secondary small RNA amplification pathway to produce 22G-RNAs (Sijen et al. 2001; Pak and Fire 2007). Additionally, RDE-1, in association with miR-243, directs the endogenous target Y47H10A.5 into the 22G-RNA pathway, leading to its silencing (Corrêa et al. 2010). To determine if endogenous 23H-RNAs also trigger 22G-RNA production, we searched sRNA-seq data sets from wild-type animals for 22 nt, 5′G-containing sequences produced from the 36 23H-RNA loci with annotated transcripts and >10 RPM 23H-RNA counts in GFP::RDE-1 co-IPs (Phillips et al. 2014). We identified 22 nt, 5′G containing small RNAs at each of these loci, and consistent with their designation as 22G-RNAs, they were almost entirely lost in mut-14 smut-1 mutants, which are defective in 22G-RNA production (Fig. 6A; Supplemental Table S5; Phillips et al. 2014). Only a small number of these loci, which included some of the highest 23H-RNA-yielding loci, such as F43E2.6, Y57G11C.1145, and Y57G11C.57, were depleted of 22G-RNAs in rde-1 mutants (Fig. 6B; Supplemental Table S5). This points to a modest or redundant role for RDE-1 in directing 22G-RNA formation from 23H-RNA targets.
Genetic requirements for 22G-RNA formation from 23H-RNA loci. (A–D) Scatter plots display individual 23H-RNA loci as the average log2-geometric mean-normalized 22G-RNA reads in wild-type (x-axis) and mutant (y-axis) animals. (A) mut-14(mg464) smut-1(tm1301). (B) rde-1(ne219). (C) prg-1(n4357). (D) ergo-1(tm1860). All libraries from gravid adults. n = 3 biological replicates for each except mut-14(mg464) smut-1(tm1301) (n = 2). (E) Average log2 ratios of 22G-RNA reads for each of the top 23H-RNA loci (>10 RPM) in the indicated mutants to wild-type. Libraries are the same as in (A–D).
Since many of the 23H-RNA loci were not substantially depleted of 22G-RNAs in rde-1 mutants, we hypothesized that additional small RNA pathways target these loci. In C. elegans, piRNAs and ERGO-1 class 26G-RNAs also direct their targets into the 22G-RNA pathway (Han et al. 2009; Gent et al. 2010; Vasale et al. 2010; Bagijn et al. 2012; Lee et al. 2012). piwi/prg-1 mutants, which lack piRNAs, produced higher levels of 22G-RNAs, based on sRNA-seq, possibly an artifact due to the loss of piRNAs and piRNA-dependent 22G-RNAs (Fig. 6C; Supplemental Table S5; Batista et al. 2008; Das et al. 2008; Wang and Reinke 2008). Only one 23H-RNA target, jmjc-1, had substantially reduced levels of 22G-RNAs in prg-1 mutants, indicating that piRNAs do not have a role in triggering 22G-RNA production from most 23H-RNA loci (Fig. 6C; Supplemental Table S5). In ergo-1 mutants, which lack oogenic/embryonic 26G-RNAs, ∼50% of the 23H-RNA loci analyzed were depleted of 22G-RNAs, which is consistent with many of these loci, such as Y17D7C.3 and K02E2.6, also being targeted by the ERGO-1 pathway (Fig. 6D; Supplemental Table S5; Han et al. 2009; Gent et al. 2010; Vasale et al. 2010; Fischer et al. 2011).
In contrast to 22G-RNAs, 23H-RNAs were not depleted in mut-14 smut-1 mutants, indicating that the Mutator pathway involved in WAGO class 22G-RNA formation is not required for 23H-RNA formation (Supplemental Fig. S7A; Supplemental Table S5). Consistent with our earlier conclusion that RDE-1 is generally dispensable for 23H-RNA formation or stability, only five individual 23H-RNAs were significantly depleted in rde-1 mutants (Supplemental Fig. S7B; Supplemental Table S5). None of the 23H-RNAs were significantly depleted in prg-1 mutants and only two were affected in ergo-1 mutants, indicating that the loss of 22G-RNAs we observed in these mutants, particularly ergo-1, is not due to the loss of 23H-RNAs (Supplemental Fig. S7C,D; Supplemental Table S5). Our results demonstrate that multiple small RNA pathways—specifically 23H-RNA, ERGO-1 class 26G-RNA, and WAGO class 22G-RNA pathways—converge on a common set of targets (Fig. 6E).
Fertility and fitness of rde-1 mutants
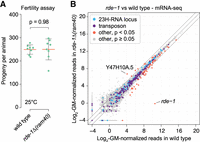
rde-1 mutants did not display any noticeable developmental defects, nor did animals with a deletion of the F43E2.6 locus, which produces the most abundant 23H-RNAs (Supplemental Fig. S8A,B). We observed a slight reduction in fertility in rde-1(ne219) mutants at 25°C, consistent with previous findings (Supplemental Fig. S8C; Seroussi et al. 2023). However, no significant reduction in progeny was observed in a distinct substitution allele of rde-1, ne300, nor in the rde-1(ram40) deletion allele (Fig. 7A; Supplemental Fig. S8C). Fertility was unchanged in F43E2.6 mutants as well (Supplemental Fig. S8C). The overall mobility and movement behavior of rde-1 and F43E2.6 mutants also did not differ significantly from wild-type animals (Supplemental Fig. S8D–F). While loss of abundant 23H-RNAs could free up bandwidth in the RNAi pathway, deletion of F43E2.6 did not enhance RNAi against the essential fertility gene hmr-1, which responds weakly to RNAi in wild-type, but can be elevated in mutants with enhanced RNAi efficacy, such as eri-5 (Supplemental Fig. S8G; Fischer et al. 2008; Thivierge et al. 2012).
Fertility and differential mRNA expression in rde-1 mutants. (A) Each data point is the number of progeny produced by a single wild-type or rde-1Δ(ram40) mutant animal grown at 25°C. The orange bars show the means and the black error bars show the 95% confidence intervals. A two-sample t-test was used to calculate the P-value. n = 8 animals per genotype. (B) The scatter plot displays individual transcripts as a function of the average log2-geometric mean-normalized reads from wild-type (x-axis) and rde-1Δ (ram40) mutant (y-axis) mRNA-seq libraries colored by classification or P-value. None of the 23H-RNA loci had P-values < 0.05. n = 3 biological replicates for each strain. Libraries from gravid adults.
We also explored the role of rde-1 in regulating endogenous genes using mRNA-seq. None of the 23H-RNA loci were differentially expressed in rde-1(ram40) mutant gravid adults, although many had too few mRNA-seq reads to evaluate (Supplemental Table S11). Despite the prevalence of 23H-RNAs derived from repetitive elements, we did not observe substantial changes to transposon expression in rde-1 mutants (Fig. 7B; Supplemental Table S11). It is possible that other small RNA pathways can compensate for the loss of rde-1 function or that alg-1 and alg-2 offset the loss of rde-1 in the 23H-RNA pathway. This could explain why rde-1 mutants lack a discernable phenotype, aside from being RNAi-defective. The loss of both alg-1 and alg-2 causes embryonic lethality, which complicates further investigation (Grishok et al. 2001).
The most significantly upregulated transcript in rde-1 mutants was Y47H10A.5, which is regulated by RDE-1 through its association with the miRNA miR-243 (Fig. 7B; Supplemental Table S11; Corrêa et al. 2010). In total, 70 transcripts were misexpressed in rde-1 mutants, which were mostly downregulated. It is unlikely that all, if any, of these are direct targets of RDE-1 since RDE-1 represses its exogenous targets via the RNAi pathway, meaning endogenous targets would be expected to be upregulated in rde-1 mutants (Fig. 7B; Supplemental Table S11). Among the differentially expressed transcripts in rde-1 mutants were 11 genes with possible roles in innate immunity, all but one of which were downregulated (GO term enrichment, P = 0.001, Benjamini correction) (Supplemental Table S11). Additionally, three genes (hsp-70, F44E5.4, and F44E5.5) encoding heat shock proteins were downregulated and two genes (vit-1 and vit-2) encoding the egg yolk precursor vitellogenin were also downregulated (Supplemental Table S11). It is possible that under less optimal growth conditions or when exposed to stress or xenobiotics, phenotypes related to these misexpressed genes could emerge in rde-1 mutants.
Discussion
In this study, we characterized the endogenous canonical siRNAs of C. elegans. We hypothesized that these siRNAs were largely overlooked in earlier studies due to their relatively low abundance compared to other small RNA classes. Our analysis confirmed this by uncovering several hundred new small RNAs from RDE-1 co-IPs that exhibit the characteristics of canonical siRNAs, many of which were rare and often undetectable in cell lysates. Our results add to the already expansive repertoire of C. elegans small RNAs and demonstrate that endogenous siRNAs like those found in mice and flies are prevalent but typically at low levels in worms. We compiled these siRNAs, along with miRNAs, 21U-RNAs, 22G-RNAs, and 26G-RNAs into a GFF3-formatted annotation file compatible with C. elegans genome releases WS235–WS290+ and WS279 annotations, which can be utilized in most high-throughput sequencing data analysis software (Supplemental Code) (updated versions will be available from MontgomeryLab.org). For optimal sensitivity, we recommend using RNA from embryos or RDE-1 co-IPs when analyzing 23H-RNAs.
RNA editing is antagonistic to RNAi and related pathways, likely because mispairs introduced by ADARs inhibit dsRNA processing by Dicer (Scadden and Smith 2001; Knight and Bass 2002; Tonkin and Bass 2003; Yang et al. 2005; Ohta et al. 2008; Heale et al. 2009; Sebastiani et al. 2009; Wu et al. 2011; Warf et al. 2012; Reich et al. 2018; Fischer and Ruvkun 2020). This could explain the rarity of most 23H-RNAs. Why some dsRNA is resistant to RNA editing and gives rise to 23H-RNAs is unclear.
It is intriguing that two of the hairpins we identified as 23H-RNA loci, mir-5592 and Y57G11C.1145, are depleted of small RNAs in pash-1 mutants, suggesting that they possess features that flag them for processing by the miRNA machinery. However, due to their strong association with RDE-1, their extended perfectly base-paired hairpins, and their tendency to produce multiple distinct small RNAs, we propose classifying them as 23H-RNAs. These loci are not conserved outside of C. elegans and may represent emerging miRNAs that have not yet been fully integrated into the miRNA pathway or regulatory networks. The differential dependence on pash-1 for hairpin-derived small RNAs is noteworthy and could indicate intermediates evolving between 23H-RNAs and miRNAs.
The hyper-enrichment of 23H-RNAs and exogenous siRNAs in RDE-1 co-IPs, compared to most miRNAs, may relate to the near-perfect double-stranded nature of the duplex intermediates. Supporting this, nearly all the most highly enriched miRNAs in GFP::RDE-1 co-IPs derive from near-perfect duplexes (<3 mismatches) (Seroussi et al. 2023). While RDE-1 shows a high affinity for 23H-RNAs, ALG-1 and ALG-2 do not appear to prefer them over miRNAs. Whether this relates to the duplex structure is unclear. It is also possible that each of these Argonautes binds small RNA duplexes produced by Dicer indiscriminately, but that RDE-1 preferentially stabilizes duplexes for which it can cleave and discard the passenger strand, a necessary step specifically in the maturation of RDE-1-small RNA complexes (Steiner et al. 2009). Our findings that 23H-RNAs are enriched in ALG-1 and ALG-2 complexes following rde-1 knockdown suggest that there is competition between the Argonautes for these small RNAs. It is possible that another factor normally assists in pairing 23H-RNAs with RDE-1, and in its absence, these siRNAs become more accessible to other Argonautes.
The mild phenotypes we observed in rde-1 mutants suggest that under optimal growth conditions, RDE-1 is not important for development. The lack of a clear phenotype in rde-1 mutants is reminiscent of the mild phenotypes originally observed in Drosophila AGO2 mutants, which appear superficially wild-type but have nonessential roles in germ cell development and male fertility, likely due to loss of siRNA activity (Okamura et al. 2004; Xu et al. 2004; Deshpande et al. 2005; Wen et al. 2015). Similar subtle phenotypes could emerge for rde-1 mutants through more detailed analyses. We identified several genes with possible roles in innate immunity downregulated in rde-1 mutants in our mRNA-seq data. It is possible that in addition to RDE-1's role in antiviral defense via the RNAi pathway, it has a role in regulating endogenous genes involved in innate immunity (Schott et al. 2005; Wilkins et al. 2005; Félix et al. 2011). Such a role could involve 23H-RNAs or miRNAs. In either case, it is possible that this or other roles for rde-1 are masked by redundancy with alg-1 and alg-2.
Future studies exploring 23H-RNAs will likely uncover their biological roles. For now, several knowledge gaps remain, including whether 23H-RNAs act in cis or trans, if they function through their association with RDE-1, ALG-1, or ALG-2, and their roles in regulating endogenous genes. Additionally, it will be important to explore their biogenesis, sorting between Argonautes, and the mechanism by which they might regulate their targets. Our study provides a framework for addressing these questions and for further exploring the 23H-RNA pathway.
Methods
Strains
USC1080[rde-1(rde-1(cmp133[(GFP + loxP + 3xFLAG)::rde-1]) V], WM27[rde-1(ne219) V], WM45[rde-1(ne300) V], PQ530[alg-1(ap423 [3xFLAG::GFP::alg-1]) X], JMC203[alg-2(tor139[GFP::3xFLAG::alg-2b]) II], JMC209[alg-5(tor145[GFP::3xFLAG::alg-5]) I], and JMC211[ergo-1(GFP::3xFLAG::ergo-1a) V] were previously described (Tabara et al. 1999; Aalto et al. 2018; Svendsen et al. 2019; Seroussi et al. 2023). TAM134[F43E2.6(ram37) II] and TAM151[rde-1(ram40) V] were generated from wild-type (N2) C. elegans using CRISPR–Cas9-mediated genome editing (Gasiunas et al. 2012; Jinek et al. 2012; Cong et al. 2013; Mali et al. 2013). Additional information is in Supplemental Material.
RNAi
Synchronized L1 larvae were grown at 20°C on E. coli HT115 expressing dsRNA matching target genes (Kamath et al. 2003). Gravid adults were collected for RNA isolation or Argonaute co-IPs at 72 h. pos-1 RNAi and associated data were from a previous experiment (Supplemental Table S12; Svendsen et al. 2019).
Protein-small RNA co-IPs
Co-IPs were performed on two or three biological replicates for each strain, as indicated. Animals were flash-frozen in liquid nitrogen and homogenized in cell lysis buffer. Cleared cell lysates were split into cell lysate and co-IP fractions. Proteins were co-IP'd with GFP-Trap Magnetic Agarose Beads (Proteintech gtma-100) for 1 h. Following three washes, beads were split between protein and RNA fractions. Protein fractions were heated at 95°C for 5 min in 1× Blue Protein Loading Dye (New England Biolabs B7703S). See Supplemental Material for additional details.
RNA isolation
RNA was isolated using TRIzol according to the manufacturer's recommendations except that two chloroform extractions were done (Invitrogen 15596018). RNA was precipitated in isopropanol overnight at −30°C in the presence of 20 μg glycogen.
Western blot
Protein fractions from GFP::3xFLAG::RDE-1 co-IPs and cell lysates were resolved on a Bolt 15-well 4%–12% Bis-Tris Plus Gel (Invitrogen NW04125BOX). Proteins were transferred to a nitrocellulose membrane and probed with FLAG (Sigma F3165) and actin (Abcam ab3280) antibodies. Bots were imaged on a FluorChem E Imaging System (ProteinSimple) and quantification was performed using ImageJ. The P-value was calculated using a two-sample t-test.
Small RNA sequencing
sRNA-seq libraries were prepared using the NEBNext Multiplex Small RNA Library Prep Set for Illumina following the manufacturer's recommendations (New England Biolabs E7300S). See Supplemental Material for additional details.
Small RNA data analysis
sRNA-seq data were analyzed and plotted using the default configuration in tinyRNA (Tate et al. 2023). For analysis of A-to-G mismatches, the tinyRNA configuration was modified to allow for up to three genome mismatches during Bowtie alignment (end_to_end: 3) and the mismatch pattern was set to ADAR (counter_mismatch_pattern: ADAR), which only counts perfect genome-matching reads and mismatched reads with 1–3 A-to-G mispairs (Langmead et al. 2009; Tate et al. 2023). Reference genome sequences and annotations were from the C. elegans WS279 genome release (Davis et al. 2022). Custom Python scripts (Supplemental Code) were used to identify complementary sequences with 2 nt, 3′-overhangs enriched in GFP::RDE-1 co-IPs from tinyRNA alignment tables generated by modifying the counter option in the configuration file (counter_diags: True). 23H-RNA and other small RNA annotations are available in GFF3 file format as Supplemental Code. Hairpin structures were predicted and drawn with RNAfold (Gruber et al. 2008). The DESeq2 R package was used within the tinyRNA pipeline to compute normalized counts using the geometric mean method and perform statistical analysis using the Wald test (Love et al. 2014). Matplotlib, R, IGV, and Adobe Illustrator were also used for plotting and statistical analysis of data (Hunter 2007; Robinson et al. 2011; R Core Team 2021). See Supplemental Table S12 for a complete list of libraries.
qRT-PCR
Small RNA TaqMan qRT-PCR was done following the manufacturer's recommendations (Invitrogen 4331348) (Supplemental Material). Two-sample t-tests were used to compare differences between conditions. Microsoft Excel and GraphPad Prism were used to draw plots and perform statistical analysis.
Microscopy
Animals were imaged 48 h (for L4 stage larvae) or 72 h (for adults) after L1 synchronization. Animals were imaged on growth media on an Axio Imager Z2 Microscope (Zeiss) using a 5X objective.
Brood size assays
Individual animals were grown on OP50 and the numbers of progeny produced by each animal were counted each day until the cessation of egg laying. For the RNAi efficacy assay, 20 animals per replicate were treated with hmr-1 RNAi, and the total progeny/replicate pool were counted at day 2 of adulthood. One-way ANOVA followed by Dunnett tests or two-sample t-tests were calculated in R or GraphPad Prism to compute P-values, as indicated (R Core Team 2021).
Behavior assays
For each assay, five 1-day-old hermaphrodites were transferred from 60 mm NGM plates with OP50 to 60 mm chemotaxis plates (CTX) (5 mM KH2PO4/K2HPO4 [pH 6.0], 1 mM CaCl2, 1 mM MgSO4, 2% agar) without food and left to roam for 2 min to adjust to the new plate. The plate was then placed on the Wormlab Imaging System (MBF Bioscience) and animal movements were recorded for 5 min (Roussel et al. 2014). Tracks were manually controlled for head and tail orientation blinded to genotype. Animals that showed no movement for 5 min or that left the field of view within 1 min were excluded from the analysis. Absolute speed (µm/s), total number of turns, and total number of reversals were captured using the Wormlab imaging system and exported to GraphPad Prism for statistical analysis. One-way ANOVA followed by a Dunnett test was used to calculate P-values.
mRNA sequencing
Total RNA from gravid adults was DNase-treated with Turbo DNase (ThermoFisher AM2238) and depleted of rRNA using Ribo-Zero (Illumina 20020596). RNA-seq libraries were prepared with the Illumina TruSeq Stranded Total RNA kit (Illumina 20020596). Samples were sequenced on an Illumina NovaSeq X Plus (PE150) by Novogene.
mRNA data analysis
Adapter trimming and quality filtering were done with fastp (Chen et al. 2018). Transcript quantification was done with RSEM, using STAR for read mapping (Li and Dewey 2011; Dobin et al. 2013). DESeq2 was used for data normalization and statistical analysis (Love et al. 2014). R was used for plotting (R Core Team 2021). Data analysis and plotting scripts are available as Supplemental Code. GO term analysis was done using the DAVID web server (Huang da et al. 2009; Sherman et al. 2022).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE254442.
Competing interest statement
The authors declare no competing interests.
Acknowledgments
Thanks to Maritza Soto-Ojeda and Reese Sprister for help with media and solutions, and Josh Svendsen and Kristen Brown for help with sRNA-seq libraries. Thanks also to Dustin Updike for the use of his laboratory while we were analyzing data. Strains were provided by the CGC, which is funded by the National Institutes of Health (P40 OD010440). This work was supported by the National Institutes of Health (R35GM119775 to T.A.M. and R01NS115947 to F.J.H.).
Footnotes
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.279083.124.
- Received February 8, 2024.
- Accepted October 3, 2024.
This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date (see https://genome.cshlp.org/site/misc/terms.xhtml). After six months, it is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.