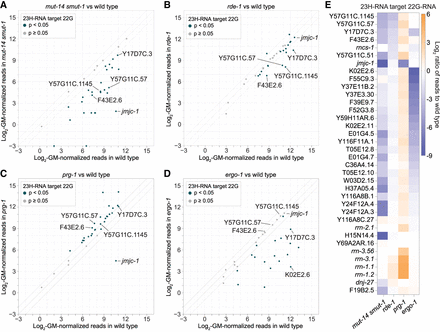
Genetic requirements for 22G-RNA formation from 23H-RNA loci. (A–D) Scatter plots display individual 23H-RNA loci as the average log2-geometric mean-normalized 22G-RNA reads in wild-type (x-axis) and mutant (y-axis) animals. (A) mut-14(mg464) smut-1(tm1301). (B) rde-1(ne219). (C) prg-1(n4357). (D) ergo-1(tm1860). All libraries from gravid adults. n = 3 biological replicates for each except mut-14(mg464) smut-1(tm1301) (n = 2). (E) Average log2 ratios of 22G-RNA reads for each of the top 23H-RNA loci (>10 RPM) in the indicated mutants to wild-type. Libraries are the same as in (A–D).











