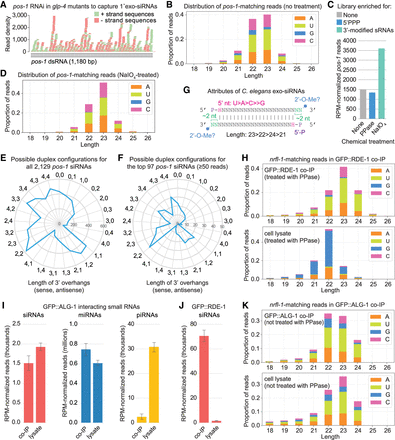
Attributes of exogenous siRNAs in C. elegans. (A) siRNA sequences matching exogenously delivered pos-1 dsRNA in glp-4(bn2) mutants as identified using sRNA-seq. n = 1 biological replicate. Libraries from adults treated with pos-1 RNAi and grown at 25°C to induce sterility. (B) Size and 5′ nt distribution of pos-1-matching reads from the sRNA-seq libraries in A. (C) RPM-normalized pos-1-matching reads from untreated (none), RNA 5′ polyphosphatase-treated (PPase, reduces 5′ triphosphates), and NaIO4-treated (blocks ligation of non-3′-end-modified small RNAs) sRNA-seq libraries. n = 1 biological replicate. RNA is the same as in the library in (A,B). (D) Size and 5′ nt distribution of pos-1-matching reads from the NaIO4-treated sRNA-seq library in C. (E) The numbers of pos-1-matching siRNA sequences with the possible duplex configurations indicated. Note that a single sequence could have multiple possible complementary strands with different 3′-overhangs. (F) As in E but limited to siRNAs with ≥50 reads. (G) Attributes of exogenous siRNAs in C. elegans. (H) Size and 5′-nt distribution of nrfl-1-matching reads from GFP::RDE-1 co-IP and cell lysate sRNA-seq libraries. n = 2 biological replicates. Data from one representative library for each condition are shown. Libraries from gravid adults treated with nrfl-1 and oma-1 RNAi. (I) RPM-normalized miRNA, piRNA, and nrfl-1-matching reads in GFP::ALG-1 co-IP and cell lysate sRNA-seq libraries. Libraries from gravid adults treated with nrfl-1 RNAi. Error bars are standard deviation (SD) between three biological replicates. (J) RPM-normalized nrfl-1-matching reads in GFP::RDE-1 co-IP and cell lysate sRNA-seq libraries. Libraries from gravid adults treated with nrfl-1 RNAi. Error bars are SD between three biological replicates. (K) Size and 5′-nt distribution of nrfl-1-matching reads from GFP::ALG-1 co-IP and cell lysate sRNA-seq libraries. Data from one representative library for each condition are shown.











