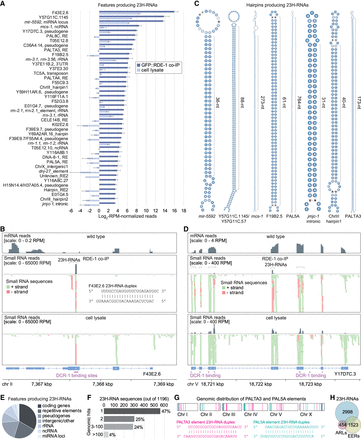
Genomic origins of 23H-RNAs. (A) Mean log2-RPM-normalized 23H-RNA reads from GFP::RDE-1 co-IP and cell lysate sRNA-seq libraries. Only features producing 23H-RNAs with >10 RPM in GFP::RDE-1 co-IPs are shown. Error bars are SD between two biological replicates. Libraries are the same as in Figure 2A. (B) mRNA-seq read density from wild-type animals and sRNA-seq read density from GFP::RDE-1 co-IP and cell lysate libraries plotted along the F43E2.6 23H-RNA locus. Brackets span 23H-RNA clusters. The lower track shows individual sequences but is truncated due to space constraints. The sequence of the most abundant duplex is shown. Purple bars span DCR-1-binding sites based on DCR-1 PAR-CLIP data from Rybak-Wolf et al. (2014). (C) Secondary structures for several 23H-RNA loci that form hairpins. (D) As in B but for the gene Y17D7C.3. (E) The proportion of 23H-RNA loci with >10 RPM in GFP::RDE-1 co-IPs overlapping the indicated features. (F) 23H-RNA sequences are categorized by the number of perfectly matching genomic loci. (G) PALTA3 and PAL5A genomic distribution and the most abundant duplex from each repetitive element. (H) Overlap between 23H-RNA loci and EERs and ARLs. Numbers shown are for total features; 449 23H-RNAs and EERs overlap; 129 23H-RNAs and ARLs overlap; 93 EERs and ARLs overlap; and 24 in common to all three data sets.











