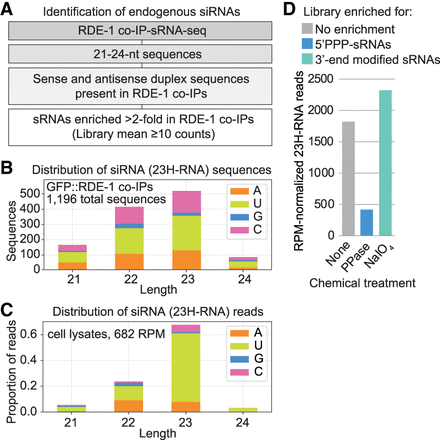
Identification of endogenous canonical siRNAs. (A) Canonical siRNA discovery flowchart. (B) Size and 5′ nt distribution of all endogenous canonical siRNA (23H-RNA) sequences from GFP::RDE-1 co-IP sRNA-seq libraries. n = 2 biological replicates. Data from one representative gravid adult library are shown. (C) Size and 5′ nt distribution of 23H-RNA reads in the cell lysates used for the GFP::RDE-1 co-IP in B. (D) 23H-RNA reads from untreated (none), RNA 5′ polyphosphatase-treated (PPase, reduces 5′ triphosphates), and NaIO4-treated (blocks ligation of non-3′-modified small RNAs) sRNA-seq libraries. n = 1 biological replicate. Libraries from wild-type gravid adults grown at 25°C.











