Spatial transcriptomics reveals asymmetric cellular responses to injury in the regenerating spiny mouse (Acomys) ear
- Henriëtte van Beijnum1,2,
- Tim Koopmans1,2,
- Antonio Tomasso1,2,
- Vanessa Disela1,2,
- Severin te Lindert1,3,
- Jeroen Bakkers1,4,
- Anna Alemany5,
- Eugene Berezikov6 and
- Kerstin Bartscherer1,2
- 1Hubrecht Institute-KNAW (Royal Netherlands Academy of Arts and Sciences), 3584CT Utrecht, The Netherlands;
- 2Department of Biology/Chemistry, Osnabrück University, 49076 Osnabrück, Germany;
- 3Wageningen University, Wageningen, 6708WE, The Netherlands;
- 4University Medical Center Utrecht, 3584CX Utrecht, The Netherlands;
- 5Department of Anatomy and Embryology, Leiden University Medical Center, and the Novo Nordisk Foundation Center for Stem Cell Medicine (reNEW), Leiden node, 2300RC Leiden, The Netherlands;
- 6European Research Institute for the Biology of Ageing, University of Groningen, University Medical Center Groningen, 9713 AV Groningen, The Netherlands
Abstract
In contrast to other mammals, the spiny mouse (Acomys) regenerates skin and ear tissue, which includes hair follicles, glands, and cartilage, in a scar-free manner. Ear punch regeneration is asymmetric with only the proximal wound side participating in regeneration. Here, we show that cues originating from the proximal side are required for normal regeneration and use spatially resolved transcriptomics (tomo-seq) to understand the molecular and cellular events underlying this process. Analyzing gene expression across the ear and comparing expression modules between proximal and distal wound sides, we identify asymmetric gene expression patterns and pinpoint regenerative processes in space and time. Moreover, using a comparative approach with nonregenerative rodents (Mus, Meriones), we strengthen a hypothesis in which particularities in the injury-induced immune response may be one of the crucial determinants for why spiny mice regenerate whereas their relatives do not. Our data are available in SpinyMine, an easy-to-use and expandable web-based tool for exploring Acomys regeneration-associated gene expression.
For decades, research in regenerative medicine has tried to find strategies to repair injuries to human tissues and organs that cannot be naturally regenerated and instead heal with scarring (Duffield et al. 2013). In contrast to humans and other adult mammals, some nonmammalian vertebrates, such as salamanders and zebrafish, and various invertebrate species, regenerate limbs and organs and have been investigated to understand the cellular and molecular basis of regeneration (Daponte et al. 2021). Although desirable because of their smaller evolutionary distance to humans and their more similar physiology, naturally regenerating mammalian model organisms are rare. Recently, a regeneration-competent mammal, the spiny mouse (Acomys spp.), has been found to show innate regenerative traits. Regeneration in Acomys was first described for the back skin and the ear pinna (Seifert et al. 2012; Gawriluk et al. 2016; Matias Santos et al. 2016) and later for skeletal muscle (Maden et al. 2018) and the spinal cord (Nogueira-Rodrigues et al. 2022). Moreover, Acomys show remarkable tolerance to ischemic injury in both the kidney (Okamura et al. 2021) and the heart (Koopmans et al. 2021; Peng et al. 2021; Qi et al. 2021) when compared to laboratory mice (Mus musculus). This makes them a promising, yet largely unexplored mammalian animal model for understanding natural tissue regeneration.
The ear pinna is the most accessible regeneration-competent tissue in Acomys and allows injury and subsequent scar-free regeneration with a mildly invasive ear punch (Gawriluk et al. 2016). The ear is a complex tissue composed of skin, which consists of epidermal keratinocytes in various differentiation stages, the underlying dermis, which hosts fibroblasts, hair follicles, and sebaceous glands, and other functional structures, such as muscle, adipose tissue, cartilage, blood vessels, and nerves, as well as immune cells. After full-thickness ear punch injury, the wound is covered by a scab of red blood cells in coagulation with a provisional extracellular matrix (ECM) and plasma proteins, followed by an inflammatory response that includes the invasion of neutrophils and monocytes, and epidermal healing with subsequent formation of a thickened and less mature wound epidermis (Gawriluk et al. 2016). This wound epidermis is thought to induce and maintain the blastema, an accumulation of cells that undergo proliferation and differentiation into the cell types needed to regenerate the lost structures (Seifert and Muneoka 2018). Although Acomys fully regenerate the ear pinna within a few weeks, most other rodents, such as Mus musculus or gerbils (Meriones unguiculatus) heal the wound with scarring, and the punch remains open. Scar formation is because of the deposition of fibrous ECM proteins, such as collagens, mainly by wound fibroblasts that populate the dermis at the wound site (Seifert et al. 2012; Gawriluk et al. 2016; Matias Santos et al. 2016).
To understand the traits that distinguish regeneration from scarring, genome-wide analyses of gene expression and protein abundance have been previously performed in Acomys versus Mus. These studies have established a correlative link between the differential expression of ECM proteins and the different responses to injury in these animals. Moreover, it was shown that differences in inflammation and the adaptive immune response exist between both species (Seifert et al. 2012; Gawriluk et al. 2016, 2020; Simkin et al. 2017a; Brewer et al. 2021). One of the remarkable aspects of ear pinna regeneration is that the missing tissue does not regenerate equally on all sides of the injury. Instead, spiny mouse ear hole closure is asymmetric with tissue growth progressing only from the proximal side of an ear punch (Matias Santos et al. 2016). This phenomenon has also been observed for small ear punches in rabbits, independent of their location along the proximal–distal axis (Williams-Boyce and Daniel 1980). Using the extraordinary regenerative abilities of spiny mice in combination with spatial transcriptomics and comparative analyses with nonregenerating rodents, we aim at identifying the molecular and cellular cues underlying asymmetric ear punch regeneration. Our data provide a comprehensive resource for the community to better understand mammalian regeneration and contribute to answering the question why some mammals regenerate whereas others do not.
Results
Quantification of asymmetric ear regeneration in Acomys
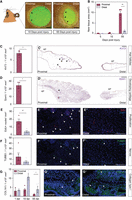
Building on the observation that ear tissue regeneration in Acomys is asymmetric and proceeds from the proximal side of the punched region towards the distal side (Matias Santos et al. 2016), we first set out to compare the extent of regeneration on both sides of the punch. We generated 4-mm ear punches in the central region of Acomys ears and quantified the new tissue from the amputation point onwards. Consistent with our previous finding that a more proximal punch regenerates faster than a distal punch (Tomasso et al. 2023), we found that a central punch regenerated relatively slowly, with new proximal tissue being visible at 10 d post ear punch injury (dpi), and >50% of the punch area still being open at 55 dpi (Fig. 1A,B). At this time point, the wound had completely healed and new tissue, including hair follicles, had formed at the proximal side of the punched area. We did not observe new hair follicles in the distal region (Fig. 1A,C). To assess the tissue composition on both sides in more detail, we quantified new cartilage, hair follicles, nerves, and differences in the number of proliferating cells in the two regions. We found Krt73+ cells, marking hair follicles, and cells arranged into a rod-like structure, presumably new cartilage, exclusively in the proximal region at 55 dpi. Cells in these cartilage-like structures were positive for mRNA expressed from the mineralization gene Sparc, a gene activated in developing cartilage in fish (Fig. 1D; Renn et al. 2006). Moreover, we detected more EdU+ cells as a measure of proliferating cells, per surface area in the proximal area compared to the distal area (Fig. 1E). In addition, we found more tubulin, beta 3 class III (TUBB3)+ cells indicating a higher density of new nerve cells in the proximal region (Fig. 1F). We also observed that the distal region showed higher levels of the fibril-forming collagen, type I, alpha 1 (COL1A1) when applying an antibody against COL1A1 on punched ear sections. This trend was apparent from 10 dpi onwards (Fig. 1G). Picrosirius red staining followed by polarized light microscopy supported this finding as it revealed an increased abundance of thick fibers and a decreased abundance of thin fibers at the distal side at 55 dpi. We detected no significant differences in the abundance of thick and thin collagen fibers between the uninjured proximal and distal regions (Supplemental Fig. S1A,B). As a major constituent of scar tissue (for review, see Keane et al. 2018), increased type I collagen levels are consistent with a regeneration-incompetent environment at the distal side of the wound.
Quantification of asymmetric ear regeneration in Acomys. (A) Regeneration of a 4-mm ear punch. Images were taken at 10 and 55 d post injury (dpi). Dotted line: original amputation plane; arrows indicate newly formed tissue. Scale bar: 1 cm. (B) Quantification of new tissue area at proximal and distal wound sites at 5, 10, and 55 dpi. N = 5. (C,D) In situ hybridization on proximal and distal ear sections (55 dpi) against Krt73 (C′) and Sparc (D′) mRNA. Barplots represent numbers of Krt73+ hair follicles (n = 3) and Sparc+ cells in the newly formed cartilage, respectively. (E) 5-ethynyl-2′-deoxyuridine (EdU) staining of proximal (E′) and distal (E′′) ear sections. n = 4. (F) Nerve cell staining and quantification (positive area) of proximal (F′) and distal (F′′) ear sections with a TUBB3 antibody. n = 5. (G) Representative images and quantifications (positive area) of proximal (G′) and distal (G′′) ear sections stained with a COL1A1 antibody. 1 dpi n = 2, 10 dpi n = 4, 55 dpi n = 3. All barplots are represented as mean ± SEM, paired t-test. Scale bar: 100 μm. Asterisks indicate significance ([∗∗] P < 0.01; [∗] P < 0.05). Arrows indicate positive staining. (See also Supplemental Fig. S1.)
Disruption of vascular and nerve supply systems inhibit regeneration
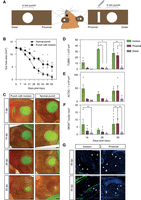
As an essential organ supply system, the vascular system extends into the body peripheries, providing oxygen, nutrients, immune cells, and hormones to even distant sites of the body. Similarly, the nervous system ensures communication between cells and tissues throughout the body and nerve axons reach far into the body peripheries. We hypothesized that an otherwise regeneration-competent distal tissue could become disconnected from systemic supply and nerve cell communication through the ear punch and consequently from any regeneration-promoting cues. Hence, we designed an experiment in which we placed an incision proximally to the ear punch. In the second ear of the same animals, we generated an ear punch without the additional incision (Fig. 2A). The presence of an incision significantly inhibited regeneration of the ear punch injury. Although the ear punch area was reduced to 4 mm2 after 4 wk in ears with only one punch, we still observed an open area of 10 mm2 in ears carrying the additional incision (Fig. 2B,C). Within 4 wk, the proximal incision had closed completely (Fig. 2C) and regeneration of the ear punch proceeded. We assessed nerve growth by quantifying the number of TUBB3+ cells in the regenerating incision and proximal tissue of the ear punch, and the nonregenerating distal side. A significant difference in the number of TUBB3+ cells between proximal and distal sides was not detected before the incision had entirely closed and a significantly higher number of TUBB3+ cells was present within the incision tissue (within 4 wk). No increase in the number of nerve cells was detected at the distal side of the punch at any time point (Fig. 2D,G). Similarly, the presence of blood vessels, as indicated by actin alpha 2, smooth muscle, aorta (ACTA2)-positive structures, was observed as early as 14 dpi in the incision region. In two out of four samples, these structures were detected in higher numbers at the proximal side than the distal side only after the incision had closed (not significant; Fig. 2E,G). Similar results were obtained with the monoclonal antibody MKI67 (also known as Ki67), as higher numbers of positive cells in the proximal blastema were only found when the incision had fully regenerated (Fig. 2F). Together, these results indicate that regeneration of the spiny mouse ear depends on regeneration-promoting cues originating from the proximal wound side, potentially provided by the unidirectional bloodstream or by nerves.
A proximal incision inhibits normal regeneration of an ear punch. (A) Schematic representation of the incision model. (B,C) Quantification of ear hole closure with and without a proximal incision (B). Representative images in C. The dotted line indicates the original amputation plane. Note that regeneration of the ear punch resumes when the proximal incision has closed. (D–F) Quantifications of incision, proximal and distal ear sections stained with a TUBB3 antibody (D), ACTA2 antibody (E), and MKI67 antibody (F). 14 dpi n = 4, 28 dpi n = 3, 55 dpi n = 4. (G) Representative images of nerve cell and vessel staining on incision tissue and proximal ear sections with TUBB3 and ACTA2 antibodies, respectively. All barplots are represented as mean ± SEM, paired t-test. Scale bar: 1 mm (C), 100 μm (G). Asterisks indicate significance ([∗∗] P < 0.01; [∗] P < 0.05). Arrows indicate positive cells.
tomo-seq determines gene expression patterns with high spatial resolution across the ear
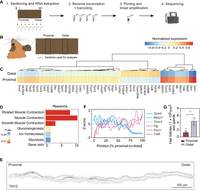
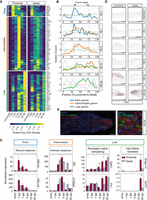
To investigate which cues from the proximal side might enable spiny mouse ear tissue to regenerate, we established spatially resolved genome-wide transcriptional profiling (tomo-seq) (Kruse et al. 2016; Wu et al. 2016; Holler and Junker 2019) for Acomys. To establish a baseline gene expression signature of uninjured ears, we sampled ear tissue from sacrificed spiny mice with a rectangular cut of 5 × 9.6 mm across the ear pinna (Fig. 3A). After snap-freezing, we processed this tissue into 96 20-μm-thick sections, from the proximal to the distal end of the rectangular tissue. Subsequent RNA extraction was performed for every fifth section, followed by reverse transcription and barcoding. The barcoded cDNA was pooled and linear-amplified, and the resulting sequencing libraries were sequenced using the Illumina NextSeq 500 (Fig. 3A). We assessed the reproducibility of the three biological replicates with the Pearson correlation coefficient between the gene expression of two different samples for all possible pairs of replicates, resulting in a correlation ranging from 0.91 to 0.99 (Supplemental Table S1), and visualized spatial profiles (Supplemental Fig. S2), demonstrating reproducibility of the replicates. To rule out the presence of baseline differences between proximal and distal areas in uninjured control tissues, we first performed a paired within-sample gene expression comparison on uninjured ears. For this analysis, the sum of the raw count data from ten sections in the middle of the proximal and distal regions was used (adjusted p adj. <0.1, log2 fold change <−1 or >1) (Fig. 3B). Differential expression and subsequent enrichment analysis showed a high abundance of muscle-related genes in the proximal compartment (Fig. 3C,D). Among these genes were myozenin 1 (Myoz1), a gene encoding a skeletal muscle Z-line protein, several troponin-coding genes (Tnnc2, Tnni2, Tnnt3), the muscle assembly regulating factor gene Tcap, and the tropomyosin-encoding genes Tpm2 and Tpm3. We visualized the distribution of muscle in the spiny mouse ear through detection of Tpm2 mRNA by in situ hybridization (Fig. 3E). Consistent with the tomo-seq data, we found muscles to be exclusively present in the proximal part of the ear (Fig. 3E,F). In addition, we found asymmetric expression of genes related to epidermal differentiation and hair follicles, such as filaggrin (Flg), desmocollin 3 (Dsc3), and cystatin E/M (Cst6) (Fig. 3C,F). These genes were enriched in the distal part of the ear, likely because of the higher density of dermal structures such as hair follicles and glands in this region (Fig. 3G). Together these data show that tomo-seq of the spiny mouse ear is highly reproducible and recapitulates the asymmetric distribution of cells and gene expression therein.
tomo-seq identifies asymmetric distribution of cell types across the Acomys ear. (A) Workflow, see text for details. Images were created with BioRender (https://www.biorender.com). (B) Schematic representation of the paired within-sample comparison on uninjured ears, proximal versus distal sections. (C) Paired within-sample comparison on uninjured ears, proximal versus distal. Shown are the top 50 differentially expressed genes (pad j <0.1, log2 fold change <−1 or >1). Color code indicates the averaged normalized expression per region (n = 2). (D) Enrichment analysis for significant genes at the proximal region using Reactome. Note that genes associated with the term “muscle” are strongly enriched in the proximal sections. (E) In situ hybridization on an ear section against Tpm2 mRNA, a muscle marker. Scale bar, 500 μm. (F) Line plot of Z-scored averaged gene expression patterns obtained by tomo-seq in the uninjured condition from the proximal to the distal side of the ear (n = 2). Shown are six representative genes selected from (C). (G) Barplot representing mean ± SEM of the counted hair follicles and glands in an uninjured ear proximal versus distal. (Paired t-test, [∗∗] P < 0.01; [∗] P < 0.05; n = 5.) (See also Supplemental Fig. S2; Supplemental Table S1.)
Asymmetric gene expression and cellular distribution in the regenerating ear
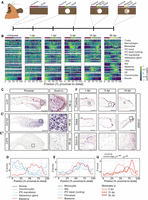
Next, we performed spatial transcriptomics on ears at early (1 h post injury [hpi], 1 dpi), intermediate (5 dpi, 10 dpi), and late stages of regeneration (55 dpi) (Fig. 4A). Cellular distribution after injury was assessed using cell type–specific marker genes (Fig. 4B; Supplemental Fig. S3A; Supplemental Table S2; Hissnauer et al. 2010; Leigh et al. 2018; Joost et al. 2020). We also used single-cell RNA-seq data from regenerating spiny mouse ears to validate the expression of the selected marker genes in the individual cell types (Supplemental Fig. S3B; Tomasso et al. 2023). Similar to the uninjured situation, we found muscle genes enriched at the proximal side of the ear punch (Fig. 4B; Supplemental Fig. S3A; Supplemental Table S2). Moreover, genes expressed primarily in chondrocytes, such as collagen, type II, alpha 1 (Col2a1) and aggrecan (Acan), were enriched in this region. Next, to assess if the tomo-seq data could recapitulate the formation of a blastema, we analyzed the expression of genes known to be activated in the limb blastema of the regeneration-competent axolotl (Leigh et al. 2018; Lin et al. 2021). From 10 dpi onwards, we detected the highest expression of these genes, such as lumican (Lum) and Twist2 (Fig. 4B–D; Supplemental Fig. S3A; Supplemental Table S2), in sections taken from the proximal wound edge, indicating the accumulation of blastema cells in this region. We found Lum not only expressed in bona fide blastema cells, but also in basal cells of the epidermis at the wound site, potentially marking the proregenerative wound epidermis (Fig. 4C).
tomo-seq identifies dynamic behavior of different cell types during ear regeneration. (A) Schematic overview of the analyzed tomo-seq samples, indicating different regions. The dotted line indicates the original amputation plane. (B) Heat map showing the average proximal–distal expression pattern of various marker genes for different cell types. n = 3 for all regeneration time points, n = 2 for uninjured samples. The white dotted lines indicate the open area. Relative higher expression in yellow and lower in blue. See Supplemental Table S2 and Supplemental Figure S3 for cell type–specific marker genes and individual line plots. (EP) Epiphyseal plate, (IFE) interfollicular epidermis, (IRS) inner root sheath, (ORS) outer root sheath. (C) In situ hybridization against blastema markers at different time points after injury: Lum (C), Col1a2 (C′), and Cd74 mRNA (C′′) on proximal ear sections. (D,E) Line plot with averaged expression patterns of cell type–specific marker genes at 55 dpi that either negatively (D) or positively correlate (E) with the expression of blastema marker genes. The dotted line indicates the open ear punch area. (F) In situ hybridization of S100a8 mRNA, a monocyte marker, at the proximal and distal wound edge at 1, 5, and 10 dpi. All scale bars, 100 μm. (G) Line plot with averaged expression patterns of marker genes for monocytes at 5, 10, and 55 dpi. The dotted line indicates the open ear punch area. (See also Supplemental Fig. S3; Supplemental Table S2.)
The expression of fibroblast-associated genes was high at the proximal side of the punch as early as 5–10 dpi and was maintained throughout the regeneration process, at least until 55 dpi. The expression of fibroblast genes, such as Col1a2, resembled those of blastema markers (Fig. 4B,C′,E; Supplemental Fig. S3A; Supplemental Table S2), consistent with the role of fibroblasts in the blastema of axolotl limbs, where they contribute to the new dermis and cartilage during limb regeneration (Lin et al. 2021). Even though mRNA levels of collagen, type I (Col1a1, Col1a2) were higher at the proximal side of the injury, likely owing to the high number of fibroblasts in the blastema, COL1A1 protein levels and thick collagen fibers were higher at the distal side (Fig. 1G; Supplemental Fig. S1). This suggests that ECM remodeling and/or collagen degradation may play a role in the ability of the proximal wound to regenerate. Next, we observed expression of genes associated with the epiphyseal plate (EP), a structure at both ends of developing long bones, characterized by the presence of hypertrophic chondrocytes, in the region of growing tissue (Fig. 4B; Supplemental Fig. S3A; Supplemental Table S2). In addition to the hypertrophic chondrocyte marker gene collagen, type X, alpha 1 encoding gene (Col10a1), we found individual cells with a chondrocyte-like shape located around the growing cartilage in the blastema at 55 dpi (Supplemental Fig. S3C). This is consistent with the occurrence of Sparc+ cells in the proximal growing ear tissue (Fig. 1D), and indicates the formation of new cartilage.
Hair follicles regenerate only at the proximal side of an Acomys ear punch wound (Figs. 1A,C, 2C). Accordingly, we detected genes known to be expressed in the inner root sheet (IRS) of developing hair follicles, such as msh homeobox 2 (Msx2) (Kulessa 2000) in proximal sections (Fig. 4B,E; Supplemental Fig. S3A; Supplemental Table S2). We also observed dynamic expression patterns of genes associated with cells of the interfollicular epidermis (IFE). Genes known to be expressed in IFE basal cells, which can proliferate to form new epidermis (Joost et al. 2020), were activated at the wound site from 5 dpi onwards. Moreover, marker genes for cycling IFE cells were enriched in the blastema region at 55 dpi (Fig. 4B,E; Supplemental Fig. S3A; Supplemental Table S2), reflecting the formation of new epidermis at the wound edge. In contrast, the expression of genes marking IFE suprabasal cells, which constitute the outer, most differentiated layers of the epidermis, and genes associated with cells of sebaceous glands and the outer root sheath (ORS), had a negative correlation with the expression of blastema markers and were comparably low in the new tissues (Fig. 4B,D; Supplemental Fig. S3A; Supplemental Table S2), indicating that the blastema contains less differentiated epidermal and dermal structures.
Next, we analyzed our data set with a focus on immune cells, as they are important for wound healing and regeneration in several animals, including spiny mice (Simkin et al. 2017a; Gaire et al. 2021). Recruitment of circulatory immune cells, such as neutrophils and monocytes from blood to extravascular sites of tissue damage, is a hallmark of early innate immune responses. By secreting cytokines, these cells attract other immune cells, such as macrophages (Ginhoux and Jung 2014; Wang et al. 2022). Consistent with the dynamic behavior of immune cells after injury, we found temporal and spatial expression differences for genes expressed in macrophages (Fig. 4B; Supplemental Fig. S3A; Supplemental Table S2). Expression of these genes appeared enriched first at the proximal blastema from 5 dpi, and then in the region surrounding the distal wound edge from 10 dpi (Fig. 4B; Supplemental Fig. S3A; Supplemental Table S2). At 55 dpi, the blastema was still positive for cells expressing CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) (Cd74) (Fig. 4C′′), indicating that macrophages are an integral part of the spiny mouse blastema also at late time points. We observed a similar shift in gene expression from the proximal wound edge at 1–5 dpi towards the distal wound edge at 10 dpi for monocyte markers, such as S100 calcium binding protein A8 (calgranulin A) (S100a8) (Fig. 4B,F,G; Supplemental Fig. S3A; Supplemental Table S2), indicating monocytes reach the proximal wound site first, with a delayed arrival at the distal wound side. In contrast to macrophage and monocyte marker genes, expression of T cell markers, such as the lymphocyte protein tyrosine kinase gene Lck, was not increased before 10 dpi. This is consistent with the late role of T cells after an injury as they are part of the adaptive immune system (Julier et al. 2017).
tomo-seq reveals distinct molecular differences between proximal and distal areas after ear punch injury
Next, we set out to identify differences in the expression of injury-induced genes between proximal and distal areas in the punched Acomys ear. The total number of reads per section for each sample was sufficient to produce a similar number of reads in all regions, including the middle sections derived from the tissue around the punch area (Supplemental Fig. S4). To avoid the possibility that the asymmetry in muscle-related genes (Fig. 3C–E) would negatively influence the identification of differentially expressed genes, we removed them from subsequent differential analyses using the Reactome muscle contraction database (R-I-397014.3) (Supplemental Table S3; Gillespie et al. 2022). However, first, we conducted a paired within-sample comparison for this gene set at each time point. This analysis resulted in an overall number of 16 differentially regulated muscle-related genes (P-value < 0.1, log2 fold change <−1 or >1), most of which showed decreasing expression in the proximal region over the regeneration process (Supplemental Fig. S5A,B). Two genes in this set, the mesenchymal marker gene vimentin (Vim) and annexin A1 (Anxa1), revealed expression patterns untypical for muscle cells (Supplemental Fig. S5B), indicating that the exclusion of the muscle gene set may have excluded genes outside of our target group.
Next, we performed differential expression analysis with the remaining transcripts for the proximal and distal areas against the corresponding uninjured control tissue for every time point. For this pairwise comparison, the raw read counts from the −5 and +5 sections surrounding the wound edge were summed (Supplemental Fig. S5C), identifying 370 differentially expressed genes (adjusted p adj. <0.1, log2 fold change <−1 or >1) in response to wounding across all time points (Supplemental Fig. S5D; Supplemental Table S4). A visual inspection of the generated heat maps indicated genes were activated or repressed in response to injury in both proximal and distal areas. Afterwards, we merged all statistically significant genes that were expressed at all time points (log2 fold change <−1 or >1 and p adj. <0.1) and clustered them according to their temporal gene expression pattern (Fig. 5A). The analysis resulted in a set of 96 differentially expressed genes, clearly indicating an elevated gene expression response in the proximal area compared to the distal area. We identified three main groups of differentially regulated genes (Fig. 5A,B): First, an early response group (“early”) was transiently induced as early as 1 hpi, containing ribosomal genes (Rps2, Rpsa) and genes involved in red blood cell maturation. Among those were genes encoding aminolevulinic acid synthase 2, erythroid (Alas2), the rate-limiting enzyme in heme biosynthesis, and putative hemoglobin subunits (Hba-a2 and Hbb-bs) (Fig. 5C). In the “early” group were also genes involved in the attraction of immune cells, such as esophageal cancer related gene 4 protein (Ecrg4), which has been associated with the recruitment of neutrophils during inflammation (Dorschner et al. 2020). The expression of the “early” group of genes was similar in proximal and distal compartments, suggesting a similar wound response at both sites of tissue damage.
tomo-seq reveals asymmetric gene expression after ear punch injury. (A) Heat map showing statistically significant genes from the differential gene expression analysis between proximal and distal areas against the corresponding uninjured control tissue (log2 fold change ≤1 or >1 and p adj. < 0.1) at 1 hpi, 1, 5, 10, and 55 dpi. Yellow indicates relative high expression; blue indicates relative low expression. Three clusters based on gene expression similarity are presented: early genes, intermediate genes, and late genes. (B) Line plot with the averaged spatial gene expression pattern of the three clusters over time; early genes (blue), intermediate genes (orange), and late genes (green). The dotted line indicates the open ear punch area. (C) Mean expression ± SEM of single genes after injury, proximal (red) and distal (light blue), for the three different clusters (n = 3 per time point). (D) In situ hybridization against Fn1 mRNA, identified in the late cluster, on proximal and distal sections. (E) Visualization of Fn1 mRNA by fluorescent in situ hybridization (red) and FN1 protein with an antibody (green) in a proximal section from an Acomys ear punch wound at 55 dpi. Nuclei are blue (DAPI). Scale bar, 100 μm. (See Supplemental Figs. S4–S7; Supplemental Tables S3, S4.)
The second group of genes, which we called the “intermediate” group, was characterized by strong expression in proximal tissues at 1–5 dpi. The intermediate group was mainly composed of genes known to be expressed in immune cells. Among those were the secretory leukocyte peptidase inhibitor (Slpi) gene, and S100a8 (Fig. 5A,C), a damage-associated molecular pattern molecule (DAMP) released from neutrophils and monocytes after tissue damage, which we had used as a marker for monocytes (Supplemental Fig. S3A). DAMP-sensing regulates transcriptional inflammatory responses in macrophages and monocytes (Austermann et al. 2022). Using in situ hybridization of Slpi and S100a8 mRNAs on sections from regenerating ear tissue, we found both genes expressed in cells close to the wound in the proximal mesenchyme during the first days after injury, highlighting the early enrichment of S100a8+ and Slpi+ immune cells at the proximal wound (Supplemental Fig. S6A). In addition, S100a8 was strongly activated in the epidermis at the wound site between 1 and 10 dpi (Fig. 4F), raising the possibility that this molecule may be involved in regulating immune cells at the wound. We also found genes in the “intermediate” group encoding attractants for monocytes, such as members of the C-C motif chemokine ligand family (Ccl2, Ccl6, and Ccl7), and C-C motif chemokine receptors (Ccr1, Ccrl2), which is consistent with the accumulation of immune cells at the wound at 1 and 5 dpi (Figs. 5B, 4F; Supplemental Fig. S6A). Despite the strong enrichment of this group in the proximal compartment, gene expression resolved more swiftly than in the distal compartment, which overall showed lower expression (Fig. 5B). Together, these data suggest that proximal and distal compartments at an ear punch injury undergo immune responses that differ from each other in strength and duration.
The third group of genes, which we called “late” genes, typically started being expressed at 5 or 10 dpi and were detected throughout the regeneration process, with highest levels measured at 55 dpi for both proximal and distal regions (Fig. 5A,B). However, the relative induction of these genes at 10 dpi was on average 1.6-fold higher in the proximal compartment, suggesting that the group of “late” genes is associated with the regeneration process and/or the presence of new structures at the site of tissue growth. Consistent with this hypothesis, we found genes expressed in hair follicles, such as Krt73, Krt25, Krtap1-4, Krtap3-2, only in the proximal tissue at 55 dpi (Fig. 5A,C; Supplemental Fig. S6A), confirming the active formation of hair follicles in the growing tissue (Fig. 1C). Prominent within the “late” group were also genes involved in the formation of the ECM, such as those encoding the transforming growth factor, beta 3(Tgfb3), a central regulator of ECM deposition, and several ECM proteins such as collagens (Col1a2, Col6a3, Col12a1) and fibronectin 1 (Fn1), which are targets of TGFB signaling in fibroblasts and other cells (Figs. 4B, 5C; Massagué 2012). Differential gene expression analysis of matrisome genes in the proximal and distal regions at different time points after injury revealed distinct matrisome profiles (Supplemental Fig. S6B,C; Supplemental Table S5). Using in situ hybridization to investigate the expression of ECM genes in more detail, we noticed the appearance of few Fn1 mRNA+ cells in the mesenchyme close to the ear punch wound in both proximal and distal compartments as early as 1 dpi. The number of Fn1+ cells increased in the following days and Fn1+ cells occupied the entire dorsal mesenchyme at the proximal, but not the distal, side of the wound. At 55 dpi, high numbers of Fn1+ mesenchymal cells were present at both proximal and distal sides, reflecting the Fn1 expression pattern observed with the tomo-seq data and confirming a delayed response of the distal side (Fig. 5C–E).
Spatial comparison with the nonregenerating rodents Mus musculus and Meriones unguiculatus
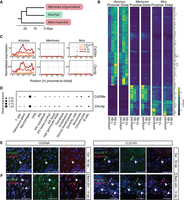
Next, we compared the Acomys injury response with that of the common laboratory mouse (Mus musculus) and the gerbil (Meriones unguiculatus) (Fig. 6A). Both animals do not regenerate 4-mm ear punches (Supplemental Fig. S7A; Gawriluk et al. 2016; Matias Santos et al. 2016). We performed tomo-seq at early (1 dpi) and intermediate (10 dpi) stages after injury, and with uninjured tissue. We figured that at the 55 dpi time point, hundreds of differentially expressed genes between Acomys and the two nonregenerators would appear because of the presence of new tissues, such as glands, cartilage, and hair follicles, in Acomys. Hence, we omitted this time point in this comparative approach. Equivalent to the Acomys data, we assessed the spatial cellular distribution after injury using the same marker genes (Supplemental Fig. S7B,C; Supplemental Table S2). We observed that like the cell distribution in Acomys, muscle and chondrocyte markers were predominantly present in the proximal part of the ear. In addition, genes marking monocytes showed a similar “shift” in gene expression from the proximal wound edge at 1 dpi to the distal wound edge at 10 dpi. We did not identify a specific enrichment of markers for macrophages in the Mus samples; however, they were enriched at the proximal wound edge at 1 dpi in Meriones. We also did not detect an accumulation of blastema markers in Mus and Meriones samples, consistent with these animals not regenerating ear tissue (Supplemental Fig. S7B,C).
Spatial comparison with the nonregenerating rodents Mus musculus and Meriones unguiculatus. (A) Phylogenetic tree showing relationships (in million years ago [mya]) between Acomys (“regenerator”), Meriones unguiculatus and Mus musculus (“nonregenerators”) (http://www.timetree.org). (B) Heat map containing two species specific clusters from the top 500 differentially expressed genes between the proximal/distal wound edge and the corresponding uninjured tissues, at 1 and 10 dpi, for Acomys, M. musculus, and M. unguiculatus. (C) Line plot with the normalized expression of Cd209a and Clec4g at the uninjured time point and at 1, 10, and 55 dpi in Acomys, M. unguiculatus, and M. musculus. (D) Dot plot visualization of gene expression of Cd209a and Clec4g in various cell types in single cell control data for Acomys. (E,F) Visualization of macrophages at the wound site of regenerating Acomys ear tissue (proximal side; 55 dpi) (E) or of regenerating Acomys back skin (14 dpi) (F) with an ADGRE1 antibody (green) and CLEC4G or CD209A (red) antibodies. Nuclei are blue (DAPI). Scale bar, 50 μm. (See Supplemental Fig. S8; Supplemental Tables S5, S6.)
Next, we investigated differences in injury-induced gene expression changes between the three animals at 1 and 10 dpi. We first compared those 96 genes we had identified as differentially expressed between proximal and distal wound sites in Acomys (Fig. 5A). We fed these genes into the same pipeline for each species individually and identified 23 genes whose expressions were less activated at the ear punch wounds of both nonregenerating animals (Supplemental Fig. S7D; Supplemental Table S4). Among those were immune cell–related genes (e.g., S100a8), ECM proteins (e.g., Mfap2, Fn1), and genes known to be involved in vascularization (e.g., Cd248, Csrp2), emphasizing the known importance of the immune system, the ECM, and angiogenesis for regeneration. Next, we performed an unbiased analysis comparing both regions and time points for all three animals. Of the top 500 differentially regulated genes (pad j < 0.1, log2 fold change < −1 or >1) (Supplemental Fig. S7E; Supplemental Table S6), 93 were more strongly activated at Acomys wounds (both proximal and distal) than at wounds of the nonregenerators, and 36 were higher expressed in the tissue of both Mus and Meriones when compared to Acomys (Fig. 6B; Supplemental Table S6).
Previous research showed a greater influx of T cells during ear pinna regeneration in Acomys and a local increase in the concentration of T cell–associated cytokines, when compared to nonregenerating mice (Gawriluk et al. 2020). Consistent with this, we found genes expressed in T cells with higher expression in Acomys. Among those was a gene encoding a member of the Interferon-induced transmembrane protein (Ifitm) family, Ifitm1, and the inhibitory fragment crystallizable receptor gene Fc receptor, IgG, low affinity IIb (Fcgr2b) (Fig. 6B; Supplemental Fig. S7F). Moreover, we detected several genes encoding members of the Lectin family of transmembrane proteins, which are known factors in the regulation of T cell behavior and function (Geijtenbeek and Gringhuis 2016). Among them was a gene encoding CD209a antigen (Cd209a; also known as DC-SIGN) and a gene encoding the C-type lectin domain family 4, member g, Clec4g. Both genes share similar temporal and spatial expression patterns, with expression starting at 1 dpi and increasing until 55 dpi in Acomys. At 55 dpi, the expression of both genes was strong at the proximal wound site (Supplemental Fig. S7G). We hardly found sequencing reads associated with these genes in the nonregenerators (Fig. 6C). In line with this, immunodetection of T cells (CD3+) and macrophages (ADGRE1+ [also known as F4/80] or CD86+), revealed increasing numbers of these immune cell types over the regeneration time course in Acomys (Supplemental Fig. S7H).
Cd209a and Clec4g are direct neighbors in a conserved cluster of structurally related lectin genes (Dominguez-Soto et al. 2007). We confirmed expression of both Clec4g and Cd209a in spiny mouse macrophages with scRNA-seq data from samples from regenerating spiny mouse ears (Tomasso et al. 2023) and with antibodies in situ (Fig. 6D; Supplemental Fig. S7G). We found CLEC4G and CD209A expression also in macrophages at the wound of injured back skin, suggesting that the presence of CLEC4G and CD209-positive macrophages is a general feature of regenerating wounds in Acomys (Fig. 6F).
Our findings reveal a specific cellular response at the proximal side of an ear punch wound that may explain the observed differences in the regenerative capacity of proximal and distal tissue. The observation that detaching the ear punch from the proximal vascular and nerve supply inhibits asymmetric regeneration (Fig. 2A–C) suggests that these injury-induced responses partially depend on cells provided by the bloodstream or the lymphatic vessels, such as CLEC4g/CD209a+ macrophages and T cells.
SpinyMine—a web-based tool for exploring Acomys tomo-seq data
To make our data accessible for the research community, we set up a freely available web tool (https://spinymine.regenerate-it.eu). This tool provides genome-wide spatial and temporal gene expression data in regenerating (Acomys) and nonregenerating rodent species (Mus musculus and Meriones unguiculatus) described in this study. Special features of this tool are spatial gene expression patterns of individual genes, comparing the expression level per region for each time point, the correlation of the gene of interest with cell type–specific markers (Supplemental Fig. S8A), comparisons between species, and links to other public databases that host further information on genes of interest.
Discussion
Here, we provide a comprehensive resource of genome-wide temporal and spatial gene expression patterns in a naturally regenerating adult mammalian tissue, the spiny mouse (Acomys) ear, and in its nonregenerative relatives, common laboratory mice (Mus) and gerbils (Meriones). Although we measured mRNA levels across the ear tissue, yielding spatially resolved individual gene expression patterns, the combination with different time points after ear punch injury enabled us to monitor the dynamic behavior of a variety of cell types during the wound healing and regeneration process in both a spatial and temporal manner. Analyzing cell type–specific markers, we uncovered the asymmetric behavior of two prominent cell populations: mesenchymal cells with a fibroblast gene expression profile and immune cells.
Accumulation of fibroblasts at the wound site
After injury, ECM is mainly deposited by fibroblasts and this ECM deposition is the major constituent of scar tissue in nonregenerating mammals (Tracy et al. 2016). Consistent with previous bulk RNA-seq and proteomics experiments (Brant et al. 2015; Gawriluk et al. 2016), we found striking differences in the expression levels of ECM genes between regenerating Acomys ears and the nonregenerating ears of Mus and Meriones. Literature showed that fibroblasts in vitro from Acomys reacted differently to induced stress than fibroblasts from Mus (Saxena et al. 2019) and that myofibroblasts in vivo after injury appeared transiently in Acomys, whereas they persisted in Mus (Brewer et al. 2021). This is consistent with studies from fish, axolotl, and mice, which showed a special role of fibroblasts and their gene expression signature in regeneration versus scarring (Currie et al. 2016; Leigh et al. 2018; Jiang and Rinkevich 2020; Lin et al. 2021; Tsata et al. 2021).
We also detected differences in the mRNA levels of ECM genes between proximal and distal wound sides, such as those of the small proteoglycan gene Lum. Lum was expressed in a subpopulation of mesenchymal cells and in basal cells of the wound epidermis. The accumulation of Lum+ cells at the site of growing tissue, and its high expression in regenerating (Acomys, Axolotl) (Leigh et al. 2018) versus scarring tissue (Mus, Meriones), may point to an important role of this ECM protein-encoding gene in providing an ECM environment favorable for regeneration and contra scarring. Lum knockout mice have a reduced wound healing capacity and thicker collagen bundles than wild-type mice (Yeh et al. 2010). Consistent with this, thick collagen fibers were enriched at the distal side, suggesting the presence of a fibrotic microenvironment that is less supportive of tissue regeneration. Our data support the idea that the proper connective tissue environment at the right time is important to allow regeneration.
Immune cells as part of the injury response
Consistent with the dynamic behavior of immune cells after injury, we found temporal and spatial differences in gene expression related to immune cells. Several genes known to be expressed in neutrophils, the population of white blood cells known to arrive first at sites of skin injury, revealed strong expression in discrete cells at the proximal wound site from 5 dpi. In light of our results from experiments in which we cut off the wound site from the blood supply, it is not surprising that immune cells arrive at the distal wound site only later. Whether the correct timing of the arrival of these cells is important for regeneration is unclear, however, their early presence may be crucial for preventing an unfavorable extracellular environment to form at the wound.
Recently it was shown that regeneration was delayed when macrophages were depleted in Acomys. However, once the ear pinna was repopulated with macrophages, regeneration was quickly restored (Simkin et al. 2017a). This is in line with previous research performed in zebrafish, mice, and axolotls, which confirmed the need for macrophages to start the regeneration process (Godwin et al. 2013; Petrie et al. 2014; Simkin et al. 2017b). Our comparative approach between Acomys and the two nonregenerators Mus and Meriones uncovered immune cell genes, which are expressed on the surface of macrophages and myeloid dendritic cells, where they are known to play a crucial role in T cell activation and differentiation (Geijtenbeek et al. 2000; Dominguez-Soto et al. 2007; Hespel and Moser 2012; Geijtenbeek and Gringhuis 2016). These findings are consistent with previous research demonstrating a greater influx of T cells during ear pinna regeneration in Acomys and a local increase in the concentration of T cell–associated cytokines when compared to nonregenerating mice (Gawriluk et al. 2020). However, the role of immune cells in regeneration is still controversial. Inflammation is a process that has been associated with scarring in nonregenerating animals, where this process lasts generally longer than in regenerators (Harty et al. 2003). Although the inflammatory response was reported to be less active in Acomys than in Mus (Brant et al. 2015), partial inhibition of inflammation by systemic inhibition of prostaglandin-endoperoxide synthase 2 (also known as cyclooxygenase 2) in Mus musculus was not sufficient to induce regeneration (Gawriluk et al. 2020). The proper timing and a fine balance between inflammatory immune responses and proregenerative functions might be important for successful regeneration.
Speed and phases of regeneration in the Acomys ear
Our study shows that structures protruding from the proximal side of the ear, such as nerve cells, muscles, or vasculature, are important for regeneration. Given a different density and architecture of nerve and vascular networks exist at proximal and distal regions in the ear, it is not surprising that regeneration of proximal punches proceeds faster than distal ones (Tomasso et al. 2023). In Acomys, it was shown (Matias Santos et al. 2016) that new axons formed only from the proximal side, raising the possibility that lacking regeneration at the distal side of an ear punch may be because of the lack of innervation. Accordingly, a study by Gawriluk et al. (2016) visualized innervation in the regenerating Acomys ear, showing that nerves sprout from the proximal side and that this side is more innervated than the distal side in both uninjured and regenerated tissues (91 dpi). In rabbits, in which asymmetric regeneration of small ear punches (2 mm) has been also observed, it has been suggested that vascularization may be responsible for the proximal–distal regeneration bias (Williams-Boyce and Daniel 1980). In line with this, our data derived from the incision model showed successful regeneration of the ear punch occurs only after a substantial number of blood vessels and nerves are present in the regenerated incision area. The accessibility of a tissue to blood, nerve, or muscle supply may therefore determine whether and how fast a tissue regenerates. The initial wave of monocyte-associated gene expression we observed first at the proximal region at 5 dpi, and only later at the distal side (Fig. 4G), supports this hypothesis.
In light of the differences in regeneration speed along the proximal–distal axis (Tomasso et al. 2023), it is desirable to dissect ear punch regeneration into distinct phases rather than reporting time points after injury alone. In our study, where we used an ear punch in the center of the wound, the ear punch was still half-way open at 55 dpi, whereas in another study, this area had already been closed at this time (Matias Santos et al. 2016). We, therefore, divided ear punch regeneration in three main phases (Supplemental Fig. S8B): An early injury response phase, starting within 1 h, is similar at both proximal and distal sides of the wound and therefore position-independent. The second phase—the “intermediate” phase—is characterized by the appearance of immune cells, in our setting between 1 and 5 dpi. This response is comparably low at the distal side. During the first days wound healing takes place: At 5 dpi, the proximal wound has been entirely covered by epidermis, whereas it is still slightly open at the distal side. In the third “late” phase, the phase of active tissue growth and differentiation of skin appendages, fibroblasts have accumulated and express high levels of ECM genes. It is likely that more phases could be discriminated when analyzing additional time points between the intermediate and the late phase. For instance, whereas we observe a group of immune cell–related genes activated transiently within the first 1–5 d after injury, we observed a second wave of immune cell–related genes, including Clec4g and Cd209a, in the growing tissue throughout the regeneration process. Whether these waves represent invading macrophages or tissue-resident cells that are stimulated to proliferate at the wound site cannot be distinguished with our tomo-seq data.
It is tempting to speculate that immune cells provided by the blood or the lymph promote regeneration at the proximal side of the wound. However, the spatial and temporal dynamics of fibroblast-associated genes and the high abundance of fibroblast-like cells at the wound site throughout the regeneration process suggest that these cells have an important role in regeneration, too, potentially by contributing a proregenerative ECM. Future research will uncover whether these cell types interact in providing a proregenerative environment at the wound site that can be eventually harvested for human regenerative therapies.
Methods
Animals
A. cahirinus, M. musculus, and M. meriones were kept in small groups in individually ventilated cages (see Supplemental Methods). All animal experiments were conducted under strict governmental and European guidelines and were approved by the Animal Welfare Committee of the Royal Netherlands Academy of Arts and Sciences, under license no. AVD80100 2018 7144.
4-mm ear punch and incision assay
Animals were anaesthetized with 4% (v/v) vaporized isoflurane and subjected to a through-and-through hole created in the middle of the right and left ear pinna using a 4-mm biopsy punch. For the incision assay, an incision proximal to the ear punch was made with a disposable knife in the right ear and the left ear was treated as described before. Following injury, animals were anaesthetized once every week and calipers were used to measure the diameter along the proximal–distal (DPD) and anterior–posterior (DAP) axes for each ear hole. Tissue was collected from sacrificed mice.
tomo-sequencing, data processing, and analysis
Ear tissue was embedded in Jung tissue freezing medium, oriented and rapidly frozen on dry ice, and stored at −80°C (as previously described in detail) (Junker et al. 2014; Kruse et al. 2016). Ear samples were cryosectioned in proximal to distal direction and once every five sections, a section with a thickness of 20 μm was collected. Total RNA was isolated using TRIzol Reagent (Invitrogen 15596018) and processed for barcoding and sequencing (see Supplemental Methods for details on sample and data processing, and analysis).
Single-cell RNA-seq data
We used the single-cell RNA-seq data by Tomasso et al. (2023) (15 dpi, Acomys control sample) for the analysis of cell type–specific gene expression (NCBI Gene Expression Ominbus [GEO; https://www.ncbi.nlm.nih.gov/geo/] accession numbers GSE224879 and GSE224433).
Immunohistochemistry
Upon tissue collection, ear samples were fixed, embedded, and processed by standard procedures. Samples were imaged using Leica SP8 microscope and Olympus VS200 slide scanner. The images were analyzed with ImageJ 1.53t. Antibodies and standard procedures are described in Supplemental Methods.
In vivo EdU labeling
Animals were injected intraperitoneally with 100-μL solution containing 1 mg/mL EdU (Invitrogen A10044) dissolved in sterile physiological saline 24 h before the point of sacrifice. EdU was visualized using the Click-iT EdU Alexa Fluor 647 imaging kit (Invitrogen C10340), according to the manufacturer's instructions. The Click-iT reaction cocktail was incubated with the samples for 1 h at RT. Tissues were then further processed according to the imaging protocol.
In situ hybridization probe synthesis
Ear tissue was snap-frozen in liquid nitrogen, and then grinded using a mortar and pestle. Total RNA was isolated, cDNA was synthesized, and probes were generated with standard procedures described in Supplemental Methods. Gene-specific primers are listed in Supplemental Table S7.
Data access
The tomo-sequencing data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA898313. tomo-seq data can be browsed in SpinyMine (https://spinymine.regenerate-it.eu).
Competing interest statement
The authors declare no competing interests.
Acknowledgments
We thank the Bayer AG and Peter Temple-Smith for providing the Acomys cahirinus founder colony, the Utrecht Sequencing Facility and Single Cell Discoveries for sequencing, Anko de Graaff and the Hubrecht Imaging Centre for assistance with microscopy, and the Hubrecht animal caretakers for spiny mouse care. We also thank the members of the Bartscherer and Bakkers groups and Steffen Werner for fruitful discussions. Research in the Bartscherer lab was supported by the Hubrecht Institute, the Osnabrueck University/Scientific Commission of Lower Saxony (WKN), the German Research Foundation (SFB1557), and the European Research Council (ERC-2016-StG 716894-IniReg). The Bartscherer and Berezikov labs are funded by the MSCA project Regenerate-IT (101073238).
Author contributions: K.B. and H.v.B. designed and oversaw the study. H.v.B. performed the bulk of the experiments and data analysis, and prepared all figures. K.B. and H.v.B. wrote the manuscript with comments from T.K. and E.B. T.K. and A.A. assisted with the data analysis. E.B. set up a database for hosting SpinyMine. S.t.L. assisted with the in situ hybridization experiments. A.T. performed the Picrosirius red experiments. V.D. performed the Acomys back skin experiments. J.B. shared protocols and expertise regarding spatial transcriptomics.
Footnotes
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.277538.122.
-
Freely available online through the Genome Research Open Access option.
- Received November 25, 2022.
- Accepted July 19, 2023.
This article, published in Genome Research, is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.