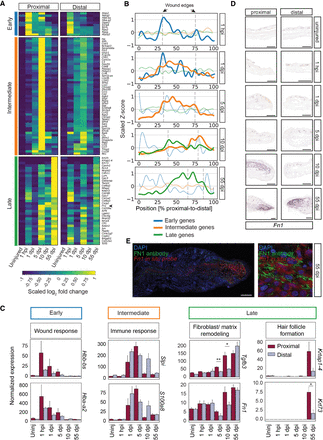
tomo-seq reveals asymmetric gene expression after ear punch injury. (A) Heat map showing statistically significant genes from the differential gene expression analysis between proximal and distal areas against the corresponding uninjured control tissue (log2 fold change ≤1 or >1 and p adj. < 0.1) at 1 hpi, 1, 5, 10, and 55 dpi. Yellow indicates relative high expression; blue indicates relative low expression. Three clusters based on gene expression similarity are presented: early genes, intermediate genes, and late genes. (B) Line plot with the averaged spatial gene expression pattern of the three clusters over time; early genes (blue), intermediate genes (orange), and late genes (green). The dotted line indicates the open ear punch area. (C) Mean expression ± SEM of single genes after injury, proximal (red) and distal (light blue), for the three different clusters (n = 3 per time point). (D) In situ hybridization against Fn1 mRNA, identified in the late cluster, on proximal and distal sections. (E) Visualization of Fn1 mRNA by fluorescent in situ hybridization (red) and FN1 protein with an antibody (green) in a proximal section from an Acomys ear punch wound at 55 dpi. Nuclei are blue (DAPI). Scale bar, 100 μm. (See Supplemental Figs. S4–S7; Supplemental Tables S3, S4.)











