A sheep pangenome reveals the spectrum of structural variations and their effects on tail phenotypes
- Ran Li1,15,
- Mian Gong1,15,
- Xinmiao Zhang1,15,
- Fei Wang1,15,
- Zhenyu Liu1,15,
- Lei Zhang1,15,
- Qimeng Yang1,
- Yuan Xu1,
- Mengsi Xu2,
- Huanhuan Zhang1,
- Yunfeng Zhang2,
- Xuelei Dai1,
- Yuanpeng Gao1,
- Zhuangbiao Zhang1,
- Wenwen Fang1,
- Yuta Yang1,
- Weiwei Fu1,
- Chunna Cao1,
- Peng Yang2,
- Zeinab Amiri Ghanatsaman3,
- Niloufar Jafarpour Negari4,
- Hojjat Asadollahpour Nanaei5,
- Xiangpeng Yue6,
- Yuxuan Song1,
- Xianyong Lan1,
- Weidong Deng7,
- Xihong Wang1,
- Chuanying Pan1,
- Ruidong Xiang8,
- Eveline M. Ibeagha-Awemu9,
- Pat (J.S.) Heslop-Harrison10,
- Benjamin D. Rosen11,
- Johannes A. Lenstra12,
- Shangquan Gan2,13 and
- Yu Jiang1,14
- 1Key Laboratory of Animal Genetics, Breeding and Reproduction of Shaanxi Province, College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi 712100, China;
- 2State Key Laboratory of Sheep Genetic Improvement and Healthy Production, Xinjiang Academy of Agricultural and Reclamation Sciences, Shihezi, Xinjiang 832000, China;
- 3Department of Animal Science, Fars Agricultural and Natural Resources Research and Education Center, Agricultural Research, Education & Extension Organization (AREEO), Shiraz 7155863511, Iran;
- 4Department of Animal Science, Shahid Bahonar University of Kerman, Kerman 7616914111, Iran;
- 5Reproductive Biotechnology Research Center, Avicenna Research Institute, ACECR, Tehran 1983969412, Iran;
- 6State Key Laboratory of Grassland Agro-ecosystems, Key Laboratory of Grassland Livestock Industry Innovation, Ministry of Agriculture and Rural Affairs, Engineering Research Center of Grassland Industry, Ministry of Education, College of Pastoral Agriculture Science and Technology, Lanzhou University, Lanzhou 730020, China;
- 7Faculty of Animal Science and Technology, Yunnan Agricultural University, Kunming 650201, China;
- 8Faculty of Veterinary & Agricultural Science, The University of Melbourne, Parkville, 3052 Victoria, Australia;
- 9Sherbrooke Research and Development Centre, Agriculture and Agri-Food Canada, Sherbrooke, Quebec J1M 0C8, Canada;
- 10Department of Genetics and Genome Biology, University of Leicester, Leicester LE1 7RH, United Kingdom;
- 11Animal Genomics and Improvement Laboratory, USDA-ARS, Beltsville, Maryland 20705, USA;
- 12Faculty of Veterinary Medicine, Utrecht University, Utrecht 3508 TD, The Netherlands;
- 13College of Coastal Agricultural Sciences, Guangdong Ocean University, Zhanjiang 524088, China;
- 14Key Laboratory of Livestock Biology, Northwest A&F University, Yangling, Shaanxi 712100, China
-
↵15 These authors contributed equally to this work.
Abstract
Structural variations (SVs) are a major contributor to genetic diversity and phenotypic variations, but their prevalence and functions in domestic animals are largely unexplored. Here we generated high-quality genome assemblies for 15 individuals from genetically diverse sheep breeds using Pacific Biosciences (PacBio) high-fidelity sequencing, discovering 130.3 Mb nonreference sequences, from which 588 genes were annotated. A total of 149,158 biallelic insertions/deletions, 6531 divergent alleles, and 14,707 multiallelic variations with precise breakpoints were discovered. The SV spectrum is characterized by an excess of derived insertions compared to deletions (94,422 vs. 33,571), suggesting recent active LINE expansions in sheep. Nearly half of the SVs display low to moderate linkage disequilibrium with surrounding single-nucleotide polymorphisms (SNPs) and most SVs cannot be tagged by SNP probes from the widely used ovine 50K SNP chip. We identified 865 population-stratified SVs including 122 SVs possibly derived in the domestication process among 690 individuals from sheep breeds worldwide. A novel 168-bp insertion in the 5′ untranslated region (5′ UTR) of HOXB13 is found at high frequency in long-tailed sheep. Further genome-wide association study and gene expression analyses suggest that this mutation is causative for the long-tail trait. In summary, we have developed a panel of high-quality de novo assemblies and present a catalog of structural variations in sheep. Our data capture abundant candidate functional variations that were previously unexplored and provide a fundamental resource for understanding trait biology in sheep.
Structural variations (SVs) are large genomic alterations of at least 50 bp. They are expected to induce larger-scale genomic perturbations in genes and regulatory regions than single-nucleotide polymorphisms (SNPs) with impact on gene expression and phenotype (Sudmant et al. 2015; Chiang et al. 2017). The spectrum of SVs has been extensively studied in the human genome in recent years, serving as a critical genetic resource for biomedical research and precision medicine (Almarri et al. 2020; Collins et al. 2020; Beyter et al. 2021; Ebert et al. 2021; Sirén et al. 2021; Wu et al. 2021). However, despite being a major source of genetic diversity (Mao et al. 2021; Leonard et al. 2022), SVs with a combination of long-read and short-read sequencing remain largely unexplored in most mammalian species including sheep.
Comprehensive characterization of SVs has been hindered by technical limitations in accurate detection and genotyping (Ebler et al. 2022), which are now being eliminated by recent advances in sequencing technologies and computational algorithms. Whereas the Pacific Biosciences (PacBio) Continuous Long Read (CLR) Mode and Nanopore long reads suffer a sequencing error rate of 5%–15% (Audano et al. 2019; Alonge et al. 2020; Liu et al. 2020; Beyter et al. 2021; Qin et al. 2021), the latest PacBio circular consensus long-read sequencing (CCS) delivers high-fidelity (HiFi) reads of 15 to 25 kb with >99.9% base accuracy (De Coster and Van Broeckhoven 2019; Wenger et al. 2019). This not only enables high-quality de novo assemblies but also empowers the resolution of SV breakpoints and sequence content with unprecedented precision and accuracy (Cheng et al. 2021; Ebert et al. 2021). Furthermore, breakthroughs in pangenome-based genome inference have largely addressed previous challenges in the discovery of diverse SV types including multiallelic complex variations (Crysnanto et al. 2021; Leonard et al. 2022). By integrating SVs and reference genome, the pangenome graph approach can particularly improve SV genotyping accuracy of short-read data (Chen et al. 2019; Ebler et al. 2022). In this way, the population frequency of otherwise inaccessible variants can be inferred in large populations made up of thousands of individuals (Sirén et al. 2021).
Sheep is among the first domesticated livestock with economic and cultural importance. Previous studies have implicated SVs in characteristics that have been under selection for millennia in sheep, including white coat color (Norris and Whan 2008), fleece variations (Demars et al. 2017), and polledness (Wiedemar and Drögemüller 2015). Nevertheless, the SV spectrum of sheep remains to be explored across populations. In this study, we take advantage of the PacBio HiFi sequencing technology to generate a diverse panel of 15 reference-quality sheep genomes and an extensive catalog of sequence-resolved SVs. We construct the first ovine pangenome and, through analysis of the SV spectrum in short sequence reads of 649 domestic and 41 wild sheep (Supplemental Table S1), highlight SVs as so far largely unexplored contributors to variations in phenotypes.
Results
High-quality de novo assemblies of 15 individuals from different breeds
We selected 15 genetically diverse individuals, including eight European (Suffolk, Dorset, Texel, Charollais, Merino, Romney, East Friesian, and Romanov), five East Asian (Ujumqin, Qiaoke, Yunnan, Waggir, and Kazakh), one Middle Eastern (Kermani), and one African (White Dorper) sheep (Fig. 1A). SNP analysis showed that the 15 individuals encompass ∼87.1% of the common SNPs (minor allele frequency [MAF] > 0.05) among 649 domestic sheep (Supplemental Fig. S1) used in this study. The 15 sheep were sequenced using PacBio HiFi sequencing at an average coverage of 20.9× (Table 1; Supplemental Table S2), yielding highly accurate long reads with average read length of 13.9 kb and per-base accuracy >99.9%. We used the hifiasm assembler (Cheng et al. 2021) for de novo assembly, which could generate one primary assembly as well as a pair of partially phased contig assemblies. The 15 primary assemblies were further anchored to chromosomal level as described below to serve as a reference panel for each breed, whereas the partially phased assemblies were used for pangenome construction and SV detection.
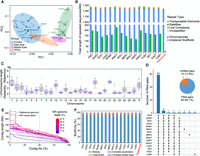
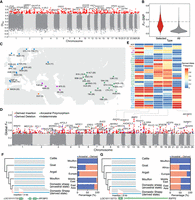
Quality assessment of 15 de novo assemblies produced by HiFi sequencing. (A) PCA plot showing the representation of the genetic diversity of domestic sheep by the 15 samples used for HiFi sequencing. The African Dorper sheep cluster with the European breeds as this breed is developed by crossing European Dorset Horn with the African Blackhead Persian sheep, resulting in its close genetic relationship with European breeds. (B) Repeat content of the 15 de novo assemblies. (C) The difference in chromosome length between the 15 new assemblies and the reference genome (REF, ARS-UI_Ram_v2.0). (D) Number of filled gaps by the different assemblies. (E) Cumulative length of contigs (Nx). In the x-axis, 50% measures the N50 across the new assemblies. HiFi sequencing depth for each assembly is shown with the dashed line referring to the reference genome. Assemblies with higher Nx values are more contiguous. (F) BUSCO completeness for the 15 new assemblies.
Quality assessments of the primary de novo assemblies for each individual and the reference genome (ARS-UI_Ramb_v2.0)
We further ordered and arranged ∼93.6% of the contigs from each primary assembly into chromosomes based on their synteny to the reference genome (ARS-UI_Ramb_v2.0). The length of the 15 primary assemblies is 2.87 Gb on average, which is closer to the estimated sheep genome size (2.84 to 3.10 Gb) (Supplemental Fig. S2; Supplemental Table S3). Most of the chromosomes are longer than their counterparts in the reference genome with a larger content of satellite sequences (Fig. 1B,C). Each assembly further possesses 135–257 Mb of the unplaced contigs that are dominated by satellite sequences (Fig. 1B) and the majority (84.0% on average) were annotated as centromeric sequences (Supplemental Table S4).
Thirteen of the 15 primary assemblies harbor fewer than 100 gaps in chromosomes (Supplemental Table S2) and the most continuous assemblies (FRI1 and DOR1) only have 28 and 35 gaps, respectively. Using whole-genome alignments where our new assemblies have non-N's sequences covering the gap region, these assemblies combined filled 82.1% (69/84) of the gaps in the reference genome (Fig. 1D). The average contig number is 1513 with a contig N50 length of 68.8 Mb. All assemblies except WDU1 have a comparable or better continuity than the reference genome (Fig. 1E). Particularly, FRI1 and DOR1 achieve contig N50 values of 85.1 and 92.4 Mb, which are much longer than that of the sheep reference genome (ARS-UI_Ramb_v2.0, 43.2 Mb). The HiFi sequencing depth is correlated positively with contig N50 (rs = 0.88) and negatively with contig number (rs = −0.62), but no effect on assembly length (rs = −0.04) was observed. In addition, the partially phased assemblies achieved a median contig N50 of 10.3 Mb, containing the haplotype-resolved alleles for each individual.
A benchmarking universal single-copy orthologs (BUSCO) analysis shows that all assemblies except WDU1 displayed a completeness that is comparable with the reference genome (Table 1; Fig. 1F). BUSCO scores for the partially phased assemblies were lower but still consistently >90% complete (Table 1; Supplemental Table S2). The quality value (QV) of the assemblies was measured using Illumina short reads sequenced to 24× on average (Table 1), which showed higher values for all the primary and phased assemblies than obtained for the reference genome (QV = 42.5), except for WDU1 (QV = 41.7).
SV discovery from PacBio HiFi reads and the ovine genome graph
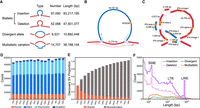
The SVs from the 15 individuals are discovered by leveraging PacBio HiFi reads and a genome graph constructed from partially phased contig assemblies (see Methods). In total, we detected 149,158 biallelic insertions/deletions (Supplemental Table S5; Supplemental Data) and abundant complex variations including 6531 divergent alleles (Supplemental Table S6) and 14,707 multiallelic variations (Supplemental Table S7; Fig. 2A). In particular, the complex variations are inferred only from the genome graph constructed by the graph constructor minigraph (see Methods), which is efficient in detecting complex variations (Leonard et al. 2022). Despite the lower contig N50 of WDU1, we did not find clear differences in the number or length distribution of detected SVs between WDU1 and other individuals (Supplemental Fig. S3). The SVs contributed 130.3 Mb of nonreference sequences in total that were dominated by TE repeats (72.5%). The insertion sequences were predicted to harbor 588 genes, whereas the deletions potentially affected the exons of 1920 genes. The nonreference sequences of the divergent alleles contain 31 genes. Among the divergent alleles, we revealed the full sequence of one divergent locus containing myeloid-associated differentiation marker (MYADM) for the first time (Fig. 2B). The MYADM locus was previously reported to be associated with mean corpuscular hemoglobin concentration and weight of lamb weaned but the sequence was unknown (Gonzalez et al. 2013). Among the multiallelic variations, we identified one region affecting interferon (IFN) gene cluster (Fig. 2C). The IFN gene clusters are key modulators of innate and adaptive immunity (Wittling et al. 2021).
Structural variation discovery in sheep. (A) Illustration and number of different SV types. The red lines indicate the reference sequence and the blue lines represent the nonreference sequence. (B) An example of a divergent allele of 50,757 bp containing a MYADM gene. (C) An example of a multiallelic variation. (D) The number of SVs per sample. (E) Pangenome growth curve generated by integrating SVs from each individual. Variants were merged starting with FRI1 followed by iteratively adding unique calls from additional samples. (F) Length distribution of different types of SVs.
As there is no “truth-set” to benchmark SV calls in sheep, we manually inspected 250 randomly selected SVs of different lengths to estimate the validation rate of detected SVs (see Methods). The overall validation rate was 94% (Supplemental Table S8). The accuracy of detected SVs was also estimated by measuring the Mendelian inheritance errors, which represent genotypes that are found in the offspring but could not be inherited from either parent. The biallelic SVs were genotyped in 11 trios of 29 individuals (average ∼18.9× of Illumina short reads) using Paragraph (Chen et al. 2019), by which 95.4% (1,609,529) of all the SVs can be genotyped. The average Mendelian error rate of 3.8% indicates a reasonable FDR based on Mendelian consistency for our SV detection and genotyping approach.
On average, 56,668 biallelic SVs were detected in each individual (Fig. 2D). The shared (present in all samples) or major (present in ≥50% of samples but not all) accounted for 63.7%, indicating that the current reference genome either carries a minor allele or an assembly error at these loci. In contrast, the polymorphic SVs and singletons account for 32.5% and 3.8%, respectively. The growth of the nonredundant set declined quickly at first and flattened when ∼10 individuals are added (Fig. 2E), which implied that a large proportion of common variants were captured by our subset of 15 sheep. The number of SVs rapidly decreases with length, consistent with reports in humans using long-read sequencing (Audano et al. 2019; Ebert et al. 2021; Quan et al. 2021). Noticeable peaks at 150 bp corresponded to short interspersed elements (SINEs), at 1300 bp to long terminal repeats (LTRs), and at 7750 bp to long interspersed nuclear elements (LINEs) (Fig. 2F; Supplemental Fig. S4). The median lengths of divergent alleles (726 bp) and multiallelic loci (575 bp) were longer than that of biallelic variations (230 bp).
As expected, the SVs are underrepresented in untranslated regions (UTRs), coding sequences (CDSs), and regulatory elements (H3K4me3, H3K27ac), suggesting the effect of negative selection (Supplemental Fig. S5). We also found 201 SV hotspots covering 182 Mb of the autosomes with enrichment in repetitive sequences (Supplemental Fig. S6). The SVs were 4.1-fold overrepresented (permutation test, P-value < 0.01) at the terminal 5 Mb region of each chromosome (Supplemental Fig. S7), which agrees with a previous study in humans (Audano et al. 2019). Furthermore, we found a 2.19-fold overrepresentation (permutation test, P-value < 0.01) of SVs in a total of 80.9 Mb (∼3.09% of the whole genome) segmental duplications (SDs), which contain 10,322 accounting for 11.4 Mb of structurally variant bases in SDs (Supplemental Fig. S8; Supplemental Tables S9, S10). The overlapped SVs (1356 bp on average) were generally shorter than the overlapped SDs (11,693 bp on average) and 9815 (95.1%) of them were entirely within SD regions, suggesting that SDs are a major driver of SVs in sheep as has been observed in other species (Bretani et al. 2020; Ebert et al. 2021).
Inference and characterization of SVs representing a derived state in sheep
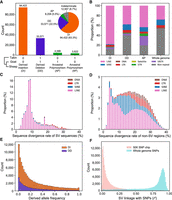
The ancestral state of a variant is critical in population genetic studies as it reflects the direction of change in an evolutionary context. The takin and goat genomes were used as outgroups for whole-genome alignments. The ancestral state of each SV was determined by estimating the size of the region in outgroups that was homologous to the region that encompassed the ovine SV (Fig. 3A). The variant state that was the same as the two outgroups was assigned as ancestral state whereas the derived-state SVs were referred to as “dSVs” as proposed by a previous study (Qin et al. 2021). The ratio of SVs’ aligned length in takin or goat to length in sheep displayed two clear peaks at 0 and 1 (Supplemental Fig. S9), corresponding to derived insertions and deletions, respectively.
Inference of derived state for SVs using takin and goat as outgroups. (A) Each SV is assigned to a derived state of either derived insertion, derived deletion, ancestral polymorphism, or indeterminate based on their presence (“1”) and absence (“0”) status in outgroup. (B) Repeat annotation in three types of dSVs. (C) Sequence divergence rate (%) of TE repeats within SV sequences. (D) Sequence divergence rate (%) of TE repeats in non-SV genomic regions. (E) dSV allele frequency spectrum. (F) Linkage analysis between SVs and nearby SNPs of whole genome (±50 kb) and ovine 50K SNP chip (±500 kb). Those SVs with MAF > 0.01 were used for linkage analysis.
The inferred dSVs included 94,422 insertions, 33,571 deletions, and 8208 putative ancestral polymorphisms (Fig. 3A). The ratio of insertions/deletions was 2.81, suggesting that insertion events likely represent the major contributor to the ovine SV spectrum. To ensure that this is not due to reference bias, we used one of our most continuous assemblies FRI1 as reference and called SVs from HiFi reads of SFK1 and still obtained much greater number of insertions than deletions (insertions/deletions: 2.46), indicating that the ovine genome indeed retained more insertions than deletions. The insertions were dominated by LINE-1 which only occupied 12.8% of the whole sheep genome but accounted for 78% of the insertions (Fig. 3B), indicating a recent LINE expansion in sheep. We further analyzed the sequence divergence rate of transposable element (TE) within SVs (Fig. 3C) and non-SV genomic regions (Fig. 3D). The LINE sequences within SVs showed lower divergence from their consensus sequences as compared with those in non-SV regions, indicating the recent active LINE expansions that contributed to a large proportion of SVs in sheep.
We genotyped all the biallelic SVs among 649 domestic sheep by Paragraph, which represents SVs as diverging paths in a graph and can achieve a high genotyping accuracy (Yan et al. 2021). The SVs are skewed toward lower frequency and the majority have a derived frequency of below 0.5 (Fig. 3E; Supplemental Table S11). Nevertheless, among the polymorphic dSVs (MAF ≥ 0.05), ∼10% were only present in domestic sheep but absent in Asiatic mouflons, and hereafter were referred to as domestication-associated SVs. In particular, 190 domestication-associated SVs were present at high frequency of >0.5 in domestic sheep, implying that they are favored by adaptation or artificial selection (Supplemental Table S12). However, it should be noted that domestication-associated SVs could also be due to genetic drift in domestication.
We further investigated how frequently the SVs were linked to nearby SNPs, seeking to estimate the extent of previously uncovered genetic variations. When using genome-wide SNP data, 59.4% of SVs showed a high linkage disequilibrium (LD) with nearby SNPs (r2 > 0.8) whereas the rest were of moderate or low LD with nearby SNPs (Fig. 3F), suggesting the presence of hidden genetic variations that cannot be captured by SNPs. Furthermore, 77.6% of the SVs displayed very low LD (r2 < 0.2) with the SNP probes from the widely used ovine 50K SNP chip, suggesting that most of the SVs could not be tagged by the SNP chip.
We found that when MAF was low (<0.05), linkage of domestication-associated SVs with nearby SNPs was significantly lower than that of standing SVs whereas when the MAF increased, the linkage of domestication-associated SVs with surrounding SNPs was enhanced and significantly stronger than that of standing SVs (Fig. 4A,B; Supplemental Figs. S10, S11). Therefore, domestication-associated SVs with high frequency tend to be subjected to selection.
The distribution of LD (r2) between SVs and nearby SNPs in domestic sheep, with different MAF ranges for SVs. (A) Density distribution of LD between SVs and nearby SNPs. (B) Contour density plots of LD between SVs and nearby SNPs. For each SV, the maximum r2 with nearby SNPs (±50 kb) on either side together with their physical distance is recorded. The Mann–Whitney U test was used to determine the difference in SV density between standing SVs and domestication-associated (Dom-associated) SVs. Blue lines: standing SVs that are present in mouflons; red lines: domestication-associated SVs.
We found the SVs were enriched with all kinds of repeats (78.3%–83.8%), whatever the SVs displaying either low, medium, or high LD with their neighboring SNPs, which were much higher than the repeat content in the whole genome (47.7%, Fig. 3B). However, the content of different repeat types of SVs from low to high LD with the nearby SNPs was different (Supplemental Fig. S12). When the LD level of SVs with nearby SNPs increased (linkage from 0–0.2 to 0.8–1), the proportion of highly repetitive regions (VNTR, STR, and Satellite) decreased from 16.0% to 1.0%, whereas the proportion of LINE increased from 51.9% to 76.0%.
Selection signatures of SVs in domestic sheep on the basis of a variant-integrated ovine pangenome
To expand our knowledge of SVs to the population scale, we explored selection signatures of SVs among 45 breeds or populations based on their SV frequency (Supplemental Table S13). The studied breeds/populations included 649 individual domestic sheep from Europe, East Asia, Middle East, and Africa, 33 Asiatic mouflons and 8 argali with an average coverage of 18.1× (Fig. 5A, see Methods for the assignment of the breeds and populations). PCA using SVs shows a clear separation of the same clusters as defined by SNPs (Supplemental Fig. S13), suggesting that the SV catalog reproduces the genetic diversity of the studied populations.
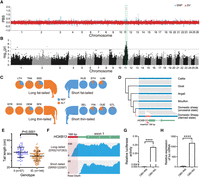
Selection signatures of SVs in domestic sheep. (A) DISV variations along the sheep genome. DISV is calculated as the derived allele frequency difference between domestic sheep and Asiatic mouflons. The top selected SVs belong to the top 1% signals from both DISV and FST-SV. (B) Distribution of the mean FST of the SNPs (FST-SNP) surrounding selected SVs as compared with all SVs in 5-kb window. The dotted line indicates the top 1% cutoff of FST-SNP distributions. (C) Geographical distribution of the 45 breeds/populations (for the breed codes, see Supplemental Table S1). The Dorper sheep from South Africa and the white Suffolk from Australia are not shown. (D) Genome-wide distribution of global FST for each SV across assigned breeds/populations. (E) The most stratified dSVs correspond to five genes associated with sheep morphology. (F) A 1.4-kb domestication-associated insertion downstream from IRF2BP2. (G) A 1.8-kb domestication-associated insertion downstream from RXFP2.
To detect SVs that are differentiated between domestics and Asiatic mouflons, we applied at each locus two differentiation indexes: DISV, which reflects the difference in derived allele frequencies between populations (Cumer et al. 2021) and FST (FST-SV). The FST-SV can identify differentiated locus, but cannot indicate the direction of selection. The DISV between domestics and wild mouflons was calculated based on derived allele frequency, allowing us to infer SVs that were under selection in domestic sheep. The intersection of 1% signals of both DISV and FST-SV were considered as the most differentiated SVs between wild and domestic populations to reduce false positives. In this way, we identified 622 SVs putatively under selection during domestication (Supplemental Table S14). A KEGG analysis of the 459 genes within 100 kb of SVs showed significant enrichment in pathways related to immunity (hepatitis B, human cytomegalovirus infection, Chagas disease, etc.) and nervous system functions (long-term depression, dopaminergic synapse, neuroactive ligand-receptor interaction, etc.) (Supplemental Table S15).
We further evaluated whether SNPs surrounding differentiated SVs were also differentiated using FST by SNPs (FST-SNP) in 5-kb window. The mean FST-SNPs around SVs were significantly higher than neutral SVs (Wilcoxon rank-sum test, P-value < 1 × 10−10). However, the surrounding FST-SNPs of most differentiated SVs (612, 98.39%) were below the 90th quantile (0.58) of the FST-SNP distributions (Fig. 5B), indicating that these SVs would probably be missed by traditional SNP-based studies. One of the plausible SVs associated with domestication is a 1.4-kb insertion in the downstream from GRIA2, of which the surrounding FST-SNPs is 0.27 (Supplemental Fig. S14). GRIA2 is a neurotransmitter in the vertebrate central nervous system and has been implicated in animal domestication (O'Rourke and Boeckx 2020).
After the initial domestication event at the Fertile Crescent, domestic sheep dispersed worldwide and adapted to a wide range of natural and artificial selection conditions. In this context, we detected putative selection signals by searching for population-stratified SVs among the 45 sheep populations using a modified global FST differentiation coefficient (Fig. 5C, see Methods). Because this strategy tends to discover common selection signals present in multiple breeds, this preferentially detected the population-stratified SVs differentiating the overrepresented European and East Asian breeds. We identified 929 population-stratified SVs with the top 1% global FST values (Fig. 5D; Supplemental Table S16), of which 352 intersect with genic regions and another 342 are within the 100-kb flanking regions of genes. The majority (725, 78.0%) were confirmed by the Ohana approach (Cheng et al. 2022), which is based on differences in allele frequencies between inferred ancestral components. The genes of the stratified SVs are associated with diverse phenotypic traits, including wool type (IRF2BP2, FGF7) (Zhu et al. 2014; Demars et al. 2017), horn type (RXFP2) (Kardos et al. 2015), fat deposition (BMP2, PDGFD, PDGFA) (Pan et al. 2019; Li et al. 2020b), and coat color (KIT, MITF) (Nishikawa et al. 1991), etc. Functional annotation of the top FST signals reveals enrichment in several KEGG pathways, such as those involved in signaling pathways (Rap1, MAPK) and parasitic disease (African trypanosomiasis, amoebiasis) (Supplemental Fig. S15).
Of the population-stratified SVs, 122 were domestication derived including 73 insertions and 49 deletions which could be key candidate variations affecting domestication and breed improvement. The highest signals are associated with five genes (IRF2BP2, BMP2, RXFP2, HOXB13, and PDGFD) (Fig. 5E) with roles in sheep morphology. The stratified dSV of IRF2BP2 is a 1.4-kb domestication-associated insertion downstream from this gene (Fig. 5F), which is reported to affect fleece variation (Demars et al. 2017). For RXFP2, the dSV is a 1.8-kb domestication-associated insertion associated with polledness (Fig. 5G; Wiedemar and Drögemüller 2015). BMP2 and PDGFD are reported to be associated with tail fat deposition in sheep (Pan et al. 2019; Li et al. 2020b; Kalds et al. 2021), whereas HOXB13 is critical in regulating vertebrate tail bud development (Denans et al. 2015). However, SVs associated with tail morphology have not been reported.
SVs as strong candidates for affecting sheep tail morphology
Tail morphology of sheep is a key genetic trait that is relevant for domestication, adaptation, productivity, and animal welfare (Kalds et al. 2021). The Asiatic mouflon has a short thin tail, whereas four domestic sheep tail types include, in addition to the primitive type, the long thin-tailed, the short fat-tailed (including fat-rumped), and the long fat-tailed types. Although several genes have been associated with these traits (Moioli et al. 2015; Xu et al. 2017; Li et al. 2020b), the underlying causative mutations are still unknown.
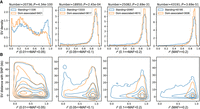
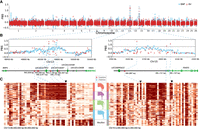
We compared SVs in the long (Asian long fat-tailed and European long thin-tailed) with those in short-tailed (Asian short fat-tailed and North European short thin-tailed) breeds within the 649 domestic sheep (Supplemental Table S1) using the population branch statistic (PBS), which is an FST-based three-population test (Yi et al. 2010) with wild mouflons as an outgroup. The most significant variation pointed to an ∼168-bp insertion (Chr 11: 37,525,005) adjacent to the annotated 5′ UTR of HOXB13 (Fig. 6A). The insertion is annotated as a SINE element with varying poly(A) sequence at the 3′ end (Supplemental Fig. S16). HOXB13 is a key regulator of caudal vertebra development. Its overexpression correlates with reduced elongation velocity in tail buds (Denans et al. 2015), whereas its repression causes extension of caudal vertebra (Economides et al. 2003; Aires et al. 2019). We measured the tail length of 201 individuals from an East Friesian × (Hu sheep × East Friesian) hybrid population and performed a genome-wide association study (GWAS) using a 40K SNP chip (see Methods) and found that the regions surrounding the 168-bp insertion are clearly associated with tail length (Fig. 6B). The insertion is prevalent in long-tailed breeds worldwide but rare in short-tailed breeds (Fig. 6C). Meanwhile, the region surrounding the insertion is highly conserved in ruminants except in sheep (Fig. 6D). This observation agrees with the short-tail status of the Caprini species. The tails of individuals homozygous for the insertion were found to be significantly longer than tails of the heterozygotes by 2.77 cm on average (Mann–Whitney U test, P-value < 0.0001) (Fig. 6E). Furthermore, the insertion was found to be expressed in RNA-seq data of colons (Clark et al. 2017), suggesting that it belongs to the 5′ UTR (Fig. 6F; Supplemental Fig. S17). To evaluate the translational efficiency, we designed plasmids containing cytomegalovirus (CMV) promoter followed by the 168-bp insertion sequence and firefly luciferase. A dual luciferase assay and a quantitative luciferase assay indicated that the insertion led to a 215-fold decrease in the luciferase protein accumulation (Fig. 6G), whereas the mRNA transcription expression decreased 29-fold (Fig. 6H). The reduction rate of transcription was not sufficient to account for the decreased luciferase expression, implying that the insertion greatly hinders protein translation.
Selective SVs associated with the long-tail trait. (A) Population branch statistic (PBS) values of long fat-tailed versus short fat-tailed sheep breeds based on dSVs and SNPs. The PBS value was calculated for SNPs using a 10-kb window size and a 5-kb step size. (B) GWAS of tail length in an East Friesian × (Hu sheep × East Friesian) hybrid population (n = 201) using a 40K SNP chip. The dotted line indicates the threshold of genome-wide significance. (C) The frequency of insertion in each breed is shown as orange in the pie chart. For the breed codes, see Supplemental Table S1. (D) The regions surrounding the insertion are highly conserved in ruminants except in sheep. (E) The carriers and noncarriers of the insertion differ in tail length. (F) RNA-seq data show the expression of the insertion in long-tailed individuals. The sequencing coverage from two RNA-seq data sets of ovine colons are shown. (G) A dual luciferase assay and a quantitative luciferase assay were used to measure the luciferase protein accumulation. The luciferase activity was measured by dual luciferase reporter assay and presented as relative LUC (firefly/Renilla luciferase). (H) Real-time PCR was used to measure the relative mRNA expression of firefly luciferase. Each experiment was repeated at least three times. Student's t-test was used to determine significance in E, G, and H. (****) P < 0.0001.
To identify candidate SVs related to the fat-tail trait, we compared the genomes of eight fat-tail breeds (SFK, KAZ, HUS, UJM, LTH, TAN, BSB, and SSS) and ten thin-tail breeds (FIN, DEM, EFR, GTL, SEL, SFK, SOG, POD, TIB, and YBB) using PBS to identify positive selection in fat-tail sheep with Asiatic mouflon as an outgroup. Two regions with the highest differentiation of either dSVs or SNPs (Fig. 7A,B) have been reported as candidate selective regions for the fat-tail trait and contain the PDGFD gene and the IBH region located between BMP2 and HAO1 (Pan et al. 2019; Baazaoui et al. 2021). We identified six highly differentiated dSVs including one domestication-associated SV in the IBH region (Fig. 7C), the largest of which is a 7728-bp insertion. PDGFD is recognized as the most plausible candidate gene for the fat-tail phenotype in recent studies (Dong et al. 2020; Li et al. 2020b). We found three highly differentiated SVs residing in the selective region of PDGFD, the largest of which is a 867-bp insertion. The three selective variants are all domestication-associated SVs. Therefore, our results suggest that both regions are associated with the fat-tail phenotype and the SVs identified within these regions are plausible candidate causative mutations.
Selective SVs associated with the fat-tail trait. (A) Population branch statistic (PBS) values across the whole genome by comparing fat-tailed sheep to thin-tailed sheep using the mouflon sheep as an outgroup. The PBS value was calculated for SNPs using a 10-kb window size and 5-kb step size. (B) The two most differentiated regions between fat-tailed sheep and thin-tailed sheep. The left panel shows the IBH region (intergenic region between BMP2 and HAO1) and the right panel corresponds to the region surrounding PDGFD. SVs with PBS > 0.8 are highlighted by the blue dotted lines. (C) The haplotypes of mouflons and domestic sheep for the two most selective regions. Each column represents one SV and each row represents one individual. The black reverted triangles represent domestication-associated SVs exclusively found in domestic sheep but absent in wild species.
Discussion
The genome-wide analysis of genomic structural variations in diverse breeds is critical to understanding the full repertoire of genetic diversity and their associations with phenotypic traits. However, previous resequencing studies mainly relied on mapping short sequence reads to a linear reference genome, which precluded the discovery of regions with pronounced structural variations (Ho et al. 2020). We performed PacBio HiFi sequencing on 15 individuals from genetically diverse breeds to generate one primary haplotype-collapsed assembly and a pair of partially phased assemblies for each breed. These assemblies provide a valuable genomic resource for in-depth functional genomic studies and captured a substantial part of the variation in Eurasian sheep. Incorporation of more individuals/breeds from Africa and America in the pangenome will enable elucidation of a more complete global variation of the sheep genome. On the other hand, our pangenome analysis is based on the reference genome. With the gradual improvement of the pangenome model (Liao et al. 2022; Wang et al. 2022b), a graph-based sheep pangenome reference will eventually be available.
Despite advances and application of third-generation sequencing technologies in human genome research (Audano et al. 2019; Almarri et al. 2020; Ebert et al. 2021; Wang et al. 2022b), long-read sequencing at the population scale is still rare for most mammalian species including sheep, which is partially due to their large genome size and limited funding of agricultural research. Therefore, the SV catalog developed in this study is the most comprehensive SV call set in mammalian species other than human and thus will deepen our knowledge of genetic diversity in sheep and other livestock species. Furthermore, we recovered a considerable number of divergent alleles and multiallelic variations by a graph-based approach using de novo assemblies, most of which are not accessible via short-read alignments. Although there is still no efficient tool to accurately genotype complex variations at the population scale, we resolved the sequences of many large variants, such as the SVs in MYADM that affects sheep erythrocyte traits (Gonzalez et al. 2013). It will be interesting to investigate the roles of other complex SVs reported in this study.
We compared the ovine SV spectrum with that of humans also generated by long-read sequencing (Ebert et al. 2021). The sheep genome harbors an average of 56,668 biallelic SVs per individual, which is more than twice the number reported (24,596 SVs per individual) in humans. We further found that the contribution of SINEs and LINEs to the SV spectrum differed between humans and sheep. The human SVs displayed a sharp peak at 300 bp for Alu, suggesting a critical role of SINEs in shaping the human SV spectrum. In contrast, the sheep genome displayed a relatively low peak at 150 bp corresponding to SINE insertions. For LINEs, sheep displayed a pronounced peak at 7.7 kb corresponding to LINE-1 insertions, and responsible for a large proportion of ovine SVs. In human, although there is a notable peak at 6.1 kb, the contribution of LINEs to human SVs is not so profound as in sheep. The longer length of LINE-1 in ruminants is likely due to their extended 5′ UTR (Boissinot and Sookdeo 2016). A marked peak at ∼8 kb for LINE-1 polymorphic insertion has also been reported in giraffe (Petersen et al. 2021), which is closer to the 7.7-kb length detected for sheep in our study. Furthermore, by inferring the ancestral state through whole-genome alignments with outgroup species, we were able to identify a larger number of derived insertions than deletions (94,422 vs. 33,571), of which LINE-1 accounted for 78% of the insertions, indicating that LINE-1 mediated insertions largely shaped the ovine SV spectrum. Considering that recent human studies mainly focused on revealing the complete SV catalog (Audano et al. 2019; Almarri et al. 2020; Ebert et al. 2021), inference of the ancestral state is warranted to provide novel insights into the origin and evolution of human SVs.
We also demonstrated how SVs could be linked to phenotypes by genotyping and assessing the SV landscape in a large cohort of data generated by short-read sequencing. We genotyped SVs using the graph-based Paragraph (Chen et al. 2019) and showed that the graph-based approach achieves high SV genotyping accuracy, which allowed us to trace natural and artificial selection responses in the studied populations. We identified a plausible causative mutation in HOXB13 for the long-tail trait, which was recently confirmed by a linkage analysis in Merinolandschaf breed (Lagler et al. 2022). Other latest research pinpointed a missense mutation in TBXT gene that could also affect tail length (Li et al. 2022). Taken together, these results suggest that sheep is a great model to study the tail development and evolution in vertebrates. Knowledge of the location of putative causative variants will enable informed precise breeding for shorter tails with the consequences of enhanced animal production performance and welfare. Therefore, SV-based association studies in larger populations accompanied with phenotypic data may very well become a powerful tool for trait discovery and application in improvement management.
In conclusion, we reported a reference panel of high-quality genome assemblies for 15 diverse sheep breeds and presented a comprehensive catalog of the SV call set. Our study is a proof-of-principle of extending the ovine pangenome to larger cohorts in order to associate SVs with more traits. The results suggest that population-scale long-read sequencing will give a new dimension to our understanding of SV prevalence and their impact on phenotypes. Logically, it may allow an effective implementation of precise genomic selection on the basis of SVs.
Methods
Sample collection and PacBio sequencing
Samples were selected to form an optimal representation of the diversity of European and Asian sheep breeds. The sampling information is provided in Supplemental Tables S2 and S17. High-quality genomic DNA (gDNA) was extracted from fresh blood samples from 15 individual sheep as previously described (Li et al. 2021) and assessed for purity and quantity using NanoDrop 1000 (Thermo Fisher Scientific) and Qubit (Thermo Fisher Scientific) assays. Libraries with an average insert size of ∼15 kb were generated using the SMRTbell Express Template Prep Kit 2.0 (PacBio) and fractionated on the SageELF (Sage Science) into narrow library fractions. Libraries were then sequenced on 2-3 SMRT Cells 8M on a Sequel II instrument (PacBio) using 30-h movie times at Annoroad Gene Technology Co., Ltd. Raw data were processed using the CCS algorithm (version 6.0.0, parameters: ‐‐minPasses 3 ‐‐minPredictedAccuracy 0.99 ‐‐maxLength 21,000) to generate highly accurate HiFi reads.
The 15 sheep were additionally sequenced to an average depth of 24× using Illumina short-read sequencing, which was used to estimate the base accuracy of de novo assemblies.
De novo assembly using HiFi reads and quality assessment
Hifiasm v0.15.3-r339 was used to generate the assembly from HiFi CCS reads using default parameters (Cheng et al. 2021). Hifiasm yields one primary contig assembly and a pair of partially phased contig assemblies. The primary contig assembly for each breed was scaffolded to chromosome level using RagTag v2.0.1 (Alonge et al. 2019) with the sheep reference genome assembly (ARS-UI_Ramb_v2.0, GCF_016772045.1).
The genome completeness was assessed using the BUSCO program v.3.0.2 containing the Mammalia odb9 set of 4104 genes (Manni et al. 2021). The single plus duplicated complete BUSCO gene counts are reported. The base accuracy was measured by assembly QV using yak (https://github.com/lh3/yak), which compares the 31-mers found in short reads and the assembly sequences.
Structural variant calling
We integrated the results from long-read-based and graph-based pangenome SV calling approach to generate high-confidence SV calls. The SVs were detected for each individual using a combination of four methods including pbsv v2.6.2 (https://github.com/PacificBiosciences/pbsv), cuteSV v1.0.13 (Jiang et al. 2020) and Sniffles v1.0.12a (Sedlazeck et al. 2018) for HiFi reads, and minigraph v0.15-r426 (Li et al. 2020a) for the partially phased assemblies.
For pbsv v2.6.2, the HiFi reads were aligned to the reference genome using pbmm2 to generate the alignment BAM file. Then the pbsv discover module was used to identify signatures of structural variation within each aligned BAM and call module to call SVs. The tandem repeat annotation file was provided to increase the sensitivity and recall of this step. For cuteSV v1.0.13, the parameters “‐‐max_cluster_bias_INS 1000 ‐‐diff_ratio_merging_INS 0.9 ‐‐max_cluster_bias_DEL 1000 ‐‐diff_ratio_merging_DEL 0.5” were used for the HiFi data. For Sniffles v1.0.12a, parameters “‐‐min_support 4 ‐‐report-seq ‐‐report_str ‐‐ccs_reads ‐‐genotype” were used.
Minigraph v0.15-r426 with option -xggs was used to integrate all partially phased contig assemblies into a graph genome (Li et al. 2020a). The sheep reference genome (ARS-UI_Ramb_v2.0) was used as the backbone of the multiassembly graph and the 30 partially phased contig assemblies were added successively according to genetic groups and ancestry (European, Middle Eastern, East Asian, African) and also by the SV numbers per individual generally in ascending order. We did not add the assemblies based on mash distance as we found the phylogenetic relationship of the two haplotypes per individual was not always consistent by the mash-based tree, potentially due to the high heterozygosity for sheep. A recent study showed that pangenomes are generally robust to different input assemblies and the order of the assemblies have little effect on the resulting pangenome (Leonard et al. 2022). Only autosomal sequences of the reference genome were used in the multigraph genome, because the X and Y Chromosomes of males are underrepresented. The SVs were called from the multiassembly graph genome using gfatools v0.5-r234.
The SVs that were identified by at least two methods were retained. We consider an insertion found by two methods if the insertion positions from the two call sets was found within 50 bp and a deletion found by two methods if they overlapped reciprocally by at least 50% using SURVIVOR v1.0.6 merge command (https://github.com/fritzsedlazeck/SURVIVOR) and BEDTools v2.29.0 (Quinlan and Hall 2010). If there was any discrepancy between the results of SV's breakpoints among the applied approaches, the breakpoints were chosen based on the preference of pbsv, cuteSV, and Sniffles.
Identification of divergent alleles and multiallelic alleles using a graph-based approach
The divergent alleles and multiallelic alleles were identified from the graph genome using a previously reported workflow (Crysnanto et al. 2021) with minor modifications. The graph generated by minigraph is composed of chains of bubbles with the reference (ARS-UI_Ramb_v2.0) as the backbone. We first used gfatools v0.5-r234, which is a bubble popping algorithm to extract bubbles from the graph (https://github.com/lh3/gfatools). Each bubble represents a structural variation, encompassing the start and end nodes of reference sequences as well as path traversing the start and end nodes.
Depending on the number of paths in a bubble, the structural variations were classified into three categories: (1) biallelic variations (insertions/deletions): either the reference path is longer (deletions) or the nonreference path is longer (insertions) and the shorter path is required to be below 100 bp. (2) Divergent alleles: the reference nodes and the nonreference nodes were both >100 bp. (3) Multiallelic alleles: one reference path and more than one nonreference path were present. Nonreference paths shorter than 100 bp were excluded.
SV validation by manual inspection and Mendelian inheritance error rate estimation
We randomly selected 250 SVs of different length with 50 SVs per length range, including 50–100 bp, 100–500 bp, 500–1000 bp, 1000–5000 bp, and >5000 bp. The SVs were manually inspected within the 15 alignment files (BAM format) from PacBio HiFi reads. Samplot (Belyeu et al. 2021) was used to check deletions and Integrative Genomics Viewer (IGV) (Thorvaldsdóttir et al. 2013) was used to identify insertions. We did not use assembly versus assembly mapping approach for SV validation, as we found that minigraph represented the variations by bubbles, which might lack precise breakpoints due to imprecise alignments in repetitive regions (Crysnanto et al. 2021).
A Mendelian inheritance error represents a genotype in the offspring that cannot be inherited from either of their parents. The whole-genome sequencing data of 11 trios were from our recent study on allelic specific expression analysis (Wang et al. 2022a). For each trio, the Mendelian error rate is the proportion of SVs from the children that deviate from Mendelian inherence.
Inferring the derived state of SV in sheep
We used high-quality genomes of takin (GCA_023091745.1) and goat (GCA_001704415.1) as an outgroup to infer the derived state of each SV based on the methods used in a previous study (Yalcin et al. 2011). First, we performed whole-genome alignment using LAST v1257 (Kiełbasa et al. 2011) to get the one-to-one pairwise alignment between genomes of sheep and two outgroup species. We used SV's flanking sequences to determine their homologous coordinates in both outgroup genomes using the command-line version of the liftOver tool (-minMatch = 0.8) from UCSC (The UCSC Genome Browser Database: update 2021). We further used BLASTN v2.10.0+ (-dust no -soft_masking false) to check the matched sequence length and similarity of the SV sequence. The presence/absence of the SV insertion in outgroup was denoted as “0” when absent in that outgroup species, “1” when present in outgroup, and “−1” when the homologous sequence could not be determined due to poor genome alignment.
According to the presence/absence status of SV sequence in outgroups, we determined the derived state of each SV (dSVs) as derived insertion (DI) if the SV was absent in two outgroups, derived deletion (DD) if present in two outgroups, ancestral polymorphism (AP) if present in one outgroup and absent in another, and indeterminate if the presence/absence could not be determined for at least one outgroup species.
SV hotspot analysis
SVs are often discovered in clusters in some specific genomic regions, which are identified as SV hotspots (Ebert et al. 2021). SV hotspot regions were determined using primatR package “hotspotter” section (bw = 200,000, num.trial = 1000, pval < 1 × 10−8) (Bakker et al. 2016). SVs were significantly clustered at the terminal 5-Mb region of each chromosome (P-value < 0.01, permutation test, random shuffling of 1000 times). The random shuffling of SV intervals was performed using BEDTools v2.29.0 with the shuffle command (Quinlan and Hall 2010).
Enrichment of SVs in the genome
The H3K4me3 and H3K27ac peak regions were retrieved from our Ruminant genome database (Fu et al. 2022). We only used insertions and deletions for enrichment and 739 SVs overlapping with centromeric or telomeric repeats were excluded. For insertions, the end position is the start position plus the insertion length. We first recorded the observed number of overlaps between SVs and UTR, exon, intron, H3K4me3, and H3K27ac peak regions, respectively. Then we counted the average number of overlaps between randomly shuffled SVs (1000 times) and each genomic region.
SV genotyping in whole-genome sequencing data
We collected whole-genome sequencing data of 649 domestic sheep, from four geographic regions (Europe, East Asia, Middle East, and Africa) as well as wild sheep including 33 Asiatic mouflons (Ovis aries musimon) and eight argali sheep (Ovis ammon) mainly from the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) (see Supplemental Table S1 for detailed information). For the domestic sheep, we selected breeds with at least five individuals and defined populations by combining breeds from the same country with less than five individuals. In this way, all the domestic sheep were assigned to 45 breeds or populations. We genotyped SVs in these samples using Paragraph v2.4a (Chen et al. 2019). The maximum permitted read count for each variant was set to 20 times the average sample depth in order to reduce runtime for repetitive regions. The resulting genotype files in VCF format from all samples were then combined using BCFtools v1.9 (Narasimhan et al. 2016).
To ensure a high-quality set of autosomal SVs for population genetics analysis, we excluded SVs that either failed to be genotyped in >90% of the samples and displayed an excess of heterozygotes deviated from Hardy–Weinberg equilibrium (P-value < 1 × 10−5) using VCFtools (Danecek et al. 2011).
The impact of sequencing coverage on genotyping efficacy was evaluated using the 15 individuals where we have both PacBio HiFi reads and Illumina reads (>20×) available. The Illumina reads were down-sampled to 2×, 5×, 10×, 15×, 20× for Paragraph genotyping to estimate the genotyping accuracy, recall, and F1 score by comparing with the genotypes from PacBio HiFi reads. In this way, the recall, accuracy, and F1 score at 5× were 0.94, 0.85, and 0.89, respectively, and with minimal increase at higher coverage (Supplemental Fig. S18).
Detecting selection signals
To detect SVs under the process of domestication, we calculated the DISV and the fixation index (FST) between wilds and domestics. The frequencies of SVs of wilds and domestics were counted using VCFtools. The FST value was calculated using the Python scikit-allel package v1.3.2 (https://github.com/cggh/scikit-allel). The FST-SNP value was calculated using the same populations and methods above. The Wilcoxon rank-sum test was carried using the Python Statistical functions in SciPy package v 0.13.0 (Virtanen et al. 2020). The 90th quantile was calculated using the Python NumPy package v 1.18.5 (Harris et al. 2020).
To detect population-stratified SVs, we employed two strategies, the global FST and the Ohana method (Cheng et al. 2022), which identifies extreme allele frequency differences by modeling ancestral admixture components. The global FST was calculated following a previously described approach with modifications (Kijas et al. 2012). For each SV site, we calculated the FST values between all populations. Instead of using the average of all FST values, we took the average of the top 50% FST values as the global FST value for each SV. The top 1% of the highest global FST value by this method were considered as putative selective signals. For the Ohana method, we first modeled the data set with the provided workflow using the SNPs (Yan et al. 2021). The resulting covariance was used as the neutral input for scans of selection. For each specific ancestry component, Ohana reported a likelihood ratio statistic (LRS) to quantify the likelihood of selection for each variant. We varied the number of ancestral admixture components from K = 2 to K = 5 to find selection signals.
In order to reveal selection signals in fat-tailed sheep, we used the method of PBS (Yi et al. 2010) to calculate and compare the FST between the fat-tailed sheep, thin-tailed sheep, and Asiatic mouflon, using the Asiatic mouflon as a distantly related population. The FST value between populations with different traits was calculated using the Python scikit-allel package v1.3.2 on the basis of both SNP and SVs. FST values for SNPs were calculated with a 10-kb sliding window with a 5-kb step.
GWAS of the tail length and PCR validation
GWAS of the tail length was conducted in an F2 population that was generated by backcrossing East Friesian sheep (♂) with the female Hu (♀) × East Friesian sheep (♂). A total of 201 individuals of similar age (6 mo old) were used. The tail length (cm) of each individual was measured and DNA samples were collected. Whole genomes were sequenced by Shijiazhuang Boruidi Biotechnology Co., Ltd. using a 40K SNP chip generated by genotyping by target sequencing (Guo et al. 2022). GWAS was conducted using GEMMA v0.98.3 with a mixed linear model (Zhou and Stephens 2012). The threshold of genome-wide significance was determined by the false discovery rate of 0.01.
The 168-bp insertion was genotyped by PCR amplification using primers binding to the flanking region of the insertion (forward: TTTATGAGCTTCTCTCCGCCA; reverse: AAGTGGTATAATTGCCGGGCT), denaturation of 94°C for 2 min followed by 28 cycles of 94°C for 30 sec, 62°C for 30 sec, and 72°C for 30 sec, an extension of 72°C for 2 min, and a hold at 4°C. PCR products were detected by agarose gel electrophoresis. As expected, the F2 population East Friesian × (Hu sheep × East Friesian) only possessed two genotypes (II and ID) due to the upgrading mating scheme.
Transfection and luciferase assays
One day before transfection, 293T cells were seeded at a density of 1 × 105 cells per well of a 24-well plate for assaying the activity of 168-bp insertion. 1.0 μg of plasmids (0.9 μg for luciferase reporter plasmid and 0.1 μg for pRL-TK) was cotransfected according to the protocol of FuGENE HD Transfection Reagent (Promega). The pRL-SV40 plasmid vector was used as an internal reference vector for standardizing transfection efficiency. Cell lysates were collected 48 h posttransfection and prepared for luciferase activity analysis using the Double-Luciferase Reporter Assay Kit (TransGen Biotec) following the manufacturer's instructions. Relative luciferase activities were expressed as the ratio of the luciferase value to the Renilla value.
RT-PCR and qPCR
Positive clones were seeded at a density of 1 × 105 cells per well of a 24-well plate. 1.0 μg of plasmids (0.9 μg for luciferase reporter plasmid and 0.1 μg for pRL-TK) was cotransfected according to the protocol of FuGENE HD Transfection Reagent (Promega). Total RNA was collected using TRIzol reagent (Invitrogen) at 48 h. Purified RNA was reverse-transcribed using a HiScript II 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme Biotech). qPCR was performed with an ABI StepOnePlus real-time PCR system (Applied Biosystems) using ChamQ SYBR qPCR Master Mix (Vazyme Biotech). The comparative Ct method was used to calculate the relative quantity of the target gene mRNA, normalized to luciferase, and was expressed as the fold change = 2-ΔΔCt. Primer sequences used for qPCR: qLuc-F: 5′-GGTGGACATTACCTACGCCGAGTA-3′; qLuc-R: CCACAGCCACACCGATGAACAG.
Data access
The data sets supporting the conclusions of this article are included within the article and its Supplemental Files. The de novo assemblies, HiFi data, and WGS data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA945429. The details are provided in Supplemental Table S17.
Competing interest statement
The authors declare no competing interests.
Acknowledgments
This study was supported by research grants from the National Key R&D Program of China (2022YFF1000100), the National Natural Science Foundation of China (U21A20247, 31822052), and the Livestock Breeding Program of Shaanxi (2022GD-TSLD-46) to Y.J., S.Q.G. (31760662, 31860639), and R.L. (31802027), Natural Science Basic Research Program of Shaanxi (2021JCW-11) to Y.J., and China Postdoctoral Science Foundation to W.W. Fu (No. 2021M702690). We thank the High-Performance Computing platform of Northwest A&F University and Hefei advanced computing center for providing computing resources. We also thank Professor X. Wang for providing the chart of different sheep tail patterns and Mr. Guochun Wang (Animal Husbandry Technology Extension Station of Chaoyang City) for help in collecting samples.
Author contributions: Y.J. and S.Q.G. conceived and designed the experiments. R.L., X.M.Z., Z.Y.L., and Q.M.Y. performed de novo assembly and SV calling; L.Z., F.W., Z.Y.L., M.S.X., and Y.F.Z. analyzed the RNA-seq data; R.L., M.G., W.W. Fang, Y.X., and X.L.D. carried out SV genotyping and population genetics analysis; Y.P.G., Y.T.Y., X.Y.L., and C.Y.P. conducted the luciferase reporter assay; Z.B.Z., W.W. Fu, H.H.Z., C.N.C., P.Y., Z.A.G., N.J.N., H.A.N., X.P.Y., Y.X.S., and W.D.D. collected the samples and prepared the WGS data; R.L. wrote the manuscript; and Y.J., S.Q.G., M.G., J.A.L., P.H.-H., E.M.I.-A., B.D.R., R.D.X., and X.H.W. performed data interpretation and revision. All authors read and approved the final manuscript.
Footnotes
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.277372.122.
-
Freely available online through the Genome Research Open Access option.
- Received September 30, 2022.
- Accepted February 21, 2023.
This article, published in Genome Research, is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.