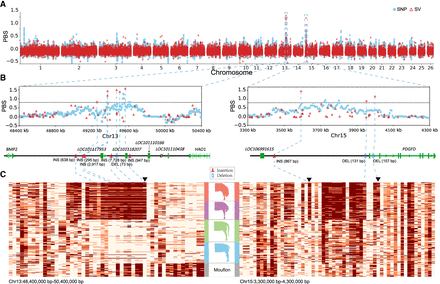
Selective SVs associated with the fat-tail trait. (A) Population branch statistic (PBS) values across the whole genome by comparing fat-tailed sheep to thin-tailed sheep using the mouflon sheep as an outgroup. The PBS value was calculated for SNPs using a 10-kb window size and 5-kb step size. (B) The two most differentiated regions between fat-tailed sheep and thin-tailed sheep. The left panel shows the IBH region (intergenic region between BMP2 and HAO1) and the right panel corresponds to the region surrounding PDGFD. SVs with PBS > 0.8 are highlighted by the blue dotted lines. (C) The haplotypes of mouflons and domestic sheep for the two most selective regions. Each column represents one SV and each row represents one individual. The black reverted triangles represent domestication-associated SVs exclusively found in domestic sheep but absent in wild species.











