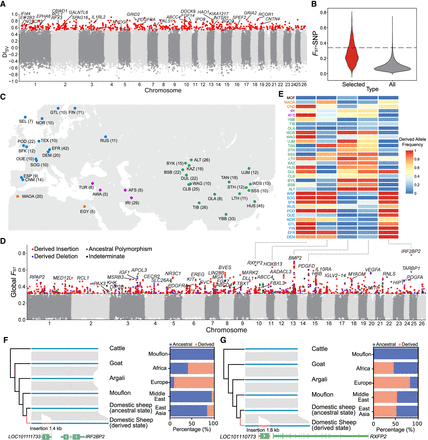
Selection signatures of SVs in domestic sheep. (A) DISV variations along the sheep genome. DISV is calculated as the derived allele frequency difference between domestic sheep and Asiatic mouflons. The top selected SVs belong to the top 1% signals from both DISV and FST-SV. (B) Distribution of the mean FST of the SNPs (FST-SNP) surrounding selected SVs as compared with all SVs in 5-kb window. The dotted line indicates the top 1% cutoff of FST-SNP distributions. (C) Geographical distribution of the 45 breeds/populations (for the breed codes, see Supplemental Table S1). The Dorper sheep from South Africa and the white Suffolk from Australia are not shown. (D) Genome-wide distribution of global FST for each SV across assigned breeds/populations. (E) The most stratified dSVs correspond to five genes associated with sheep morphology. (F) A 1.4-kb domestication-associated insertion downstream from IRF2BP2. (G) A 1.8-kb domestication-associated insertion downstream from RXFP2.











