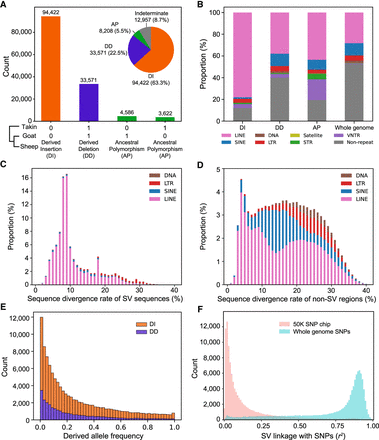
Inference of derived state for SVs using takin and goat as outgroups. (A) Each SV is assigned to a derived state of either derived insertion, derived deletion, ancestral polymorphism, or indeterminate based on their presence (“1”) and absence (“0”) status in outgroup. (B) Repeat annotation in three types of dSVs. (C) Sequence divergence rate (%) of TE repeats within SV sequences. (D) Sequence divergence rate (%) of TE repeats in non-SV genomic regions. (E) dSV allele frequency spectrum. (F) Linkage analysis between SVs and nearby SNPs of whole genome (±50 kb) and ovine 50K SNP chip (±500 kb). Those SVs with MAF > 0.01 were used for linkage analysis.











