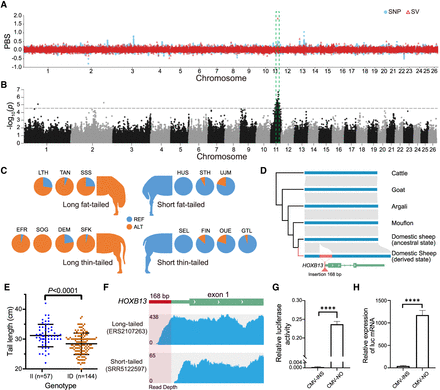
Selective SVs associated with the long-tail trait. (A) Population branch statistic (PBS) values of long fat-tailed versus short fat-tailed sheep breeds based on dSVs and SNPs. The PBS value was calculated for SNPs using a 10-kb window size and a 5-kb step size. (B) GWAS of tail length in an East Friesian × (Hu sheep × East Friesian) hybrid population (n = 201) using a 40K SNP chip. The dotted line indicates the threshold of genome-wide significance. (C) The frequency of insertion in each breed is shown as orange in the pie chart. For the breed codes, see Supplemental Table S1. (D) The regions surrounding the insertion are highly conserved in ruminants except in sheep. (E) The carriers and noncarriers of the insertion differ in tail length. (F) RNA-seq data show the expression of the insertion in long-tailed individuals. The sequencing coverage from two RNA-seq data sets of ovine colons are shown. (G) A dual luciferase assay and a quantitative luciferase assay were used to measure the luciferase protein accumulation. The luciferase activity was measured by dual luciferase reporter assay and presented as relative LUC (firefly/Renilla luciferase). (H) Real-time PCR was used to measure the relative mRNA expression of firefly luciferase. Each experiment was repeated at least three times. Student's t-test was used to determine significance in E, G, and H. (****) P < 0.0001.











