Unique genetic basis of the distinct antibiotic potency of high acetic acid production in the probiotic yeast Saccharomyces cerevisiae var. boulardii
- Benjamin Offei1,2,3,
- Paul Vandecruys1,2,3,
- Stijn De Graeve1,2,
- María R. Foulquié-Moreno1,2 and
- Johan M. Thevelein1,2
- 1Laboratory of Molecular Cell Biology, Institute of Botany and Microbiology, KU Leuven, B-3001 Leuven-Heverlee, Flanders, Belgium;
- 2Center for Microbiology, VIB, B-3001 Leuven-Heverlee, Flanders, Belgium
-
↵3 These authors contributed equally to this work.
Abstract
The yeast Saccharomyces boulardii has been used worldwide as a popular, commercial probiotic, but the basis of its probiotic action remains obscure. It is considered conspecific with budding yeast Saccharomyces cerevisiae, which is generally used in classical food applications. They have an almost identical genome sequence, making the genetic basis of probiotic potency in S. boulardii puzzling. We now show that S. boulardii produces at 37°C unusually high levels of acetic acid, which is strongly inhibitory to bacterial growth in agar-well diffusion assays and could be vital for its unique application as a probiotic among yeasts. Using pooled-segregant whole-genome sequence analysis with S. boulardii and S. cerevisiae parent strains, we succeeded in mapping the underlying QTLs and identified mutant alleles of SDH1 and WHI2 as the causative alleles. Both genes contain a SNP unique to S. boulardii (sdh1F317Y and whi2S287*) and are fully responsible for its high acetic acid production. S. boulardii strains show different levels of acetic acid production, depending on the copy number of the whi2S287* allele. Our results offer the first molecular explanation as to why S. boulardii could exert probiotic action as opposed to S. cerevisiae. They reveal for the first time the molecular-genetic basis of a probiotic action-related trait in S. boulardii and show that antibacterial potency of a probiotic microorganism can be due to strain-specific mutations within the same species. We suggest that acquisition of antibacterial activity through medium acidification offered a selective advantage to S. boulardii in its ecological niche and for its application as a probiotic.
Some microorganisms have found beneficial applications as probiotics in humans (Macfarlane and Cummings 2002; Senok et al. 2009; Butel 2014; Szajewska 2016), animal husbandry (Chaucheyras-Durand and Durand 2010; Hou et al. 2015), and aquaculture (Verschuere et al. 2000; Balcazar et al. 2006). Probiotics are live microorganisms that confer a health benefit to the host. They consist mainly of bacterial strains and also one specific yeast strain Saccharomyces cerevisiae var. boulardii (S. boulardii) (Elmer et al. 1996; Guandalini 2011). Clinical evidence abounds for the beneficial applications of probiotics in the treatment of multiple gastrointestinal disorders (Guandalini 2008, 2011; McFarland 2009). This is also true for the yeast S. boulardii (McFarland 2010; Girardin and Seidman 2011; Currò et al. 2017). It is the only yeast strain that is prescribed as a probiotic against gastrointestinal diseases and is commercially available from pharmacies worldwide. Studies in animal models and clinical trials in patients have shown its effectiveness against many gut-related diseases, including Crohn's disease (McFarland 2010), ulcerative colitis (Guslandi et al. 2003), antibiotic-associated diarrhea (McFarland et al. 1995; Duman et al. 2005; Kotowska et al. 2005), gut inflammatory manifestations in HIV patients (Villar-García et al. 2015), and recurrent Clostridium difficile infections (McFarland et al. 1994; McFarland 2009). S. boulardii is also known to ameliorate diarrhea as a result of gastrointestinal infections by bacterial enteropathogens (Czerucka et al. 2007). Certain strains of S. cerevisiae also appear to have probiotic capacity (Martins et al. 2007; Zanello et al. 2011a,b), but protection against enteric bacterial pathogens has not been reported. Their phylogenetic relationship with S. boulardii is not known.
The origin of S. boulardii can be traced back to southeast Asia, where it was first isolated from lychee and mangosteen fruits in 1920 by Henry Boulard (McFarland 2010). Although previously considered a different species, modern molecular phylogenetic methods suggest that it is a variety of the baker's yeast, Saccharomyces cerevisiae (McCullough et al. 1998; Mitterdorfer et al. 2002; van der Aa Kühle and Jespersen 2003; Edwards-Ingram et al. 2004; Posteraro et al. 2005; MacKenzie et al. 2008). More recent whole-genome sequence analysis has revealed that S. boulardii shares a highly similar genome sequence with S. cerevisiae (Khatri et al. 2013, 2017). In spite of this, S. boulardii exhibits several distinct metabolic and physiological characteristics. It shows much better tolerance to acidic conditions akin to that of the gastric milieu (Fietto et al. 2004; Edwards-Ingram et al. 2007; Cascio et al. 2013), possesses enhanced ability for pseudohyphal switching (Edwards-Ingram et al. 2007), and thrives better at 37°C (Fietto et al. 2004). It has remained unclear as to how far these distinct properties are important for its probiotic potency.
Multiple mechanisms have been proposed for the probiotic action of S. boulardii. These include modulation of cytokine production (Dalmasso et al. 2006; Mumy et al. 2008), stimulation of immunoglobulin A production against C. difficile toxin A (Qamar et al. 2001), and degradation of the toxin and host toxin receptors by a secreted protease (Pothoulakis et al. 1993; Castagliuolo et al. 1996). S. boulardii may preserve enterocyte barrier integrity by stimulating tight junction protein secretion and may exclude bacterial pathogens from interaction with intestinal epithelial cells by directly binding to the pathogens (Gedek 1999; Martins et al. 2010; Tiago et al. 2012).
Secretion of antimicrobial compounds in the form of peptides (bacteriocins), hydrogen peroxide, or organic acids features prominently among the generally accepted action mechanisms of bacterial probiotics (Cursino et al. 2006; Hutt et al. 2006; Pridmore et al. 2008; Girardin and Seidman 2011; Ciorba 2012; Dobson et al. 2012; O'Shea et al. 2012; Tejero-Sariñena et al. 2012; Patel and DuPont 2015; Lopes et al. 2017). Direct inhibitory action on bacterial growth or antimicrobial compound secretion by S. boulardii has never been reported.
As opposed to most S. cerevisiae strains, S. boulardii lacks the ability to sporulate, which has severely hampered genetic analysis of its specific attributes, especially those that may be responsible for its probiotic potency (McCullough et al. 1998; van der Aa Kühle and Jespersen 2003; Edwards-Ingram et al. 2007). As a result, the genetic differences between S. boulardii and S. cerevisiae that might explain the superior probiotic potency of the former have remained completely unknown. On the other hand, genetic linkage studies, such as pooled-segregant whole-genome sequence analysis, combined with reciprocal hemizygosity analysis (RHA) and allele exchange for identifying causative alleles and SNPs, have proven very effective in dissecting the polygenic basis of commercially important traits in different S. cerevisiae strains (Liti and Louis 2012; Swinnen et al. 2012).
The aim of this study was to elucidate the genetic basis of the unusually high acetic acid levels we found to be produced by S. boulardii, to evaluate their antibacterial potency as a possible explanation for the probiotic action of S. boulardii, and to assess whether this genetic basis could serve as a specific genetic signature for distinguishing S. boulardii and S. cerevisiae.
Results
Classification of S. boulardii and S. cerevisiae strains using amplified fragment length polymorphisms (AFLPs) and whole-genome sequence analysis
S. boulardii strains obtained from various sources (Supplemental Table S1) were characterized alongside 23 S. cerevisiae strains, one S. mikatae, and one S. paradoxus strain, using amplified fragment length polymorphisms (AFLPs) and whole-genome sequence analysis. All S. boulardii strains formed a single cluster of highly related strains (Supplemental Fig. S1). Furthermore, this cluster of S. boulardii strains was embedded within a larger S. cerevisiae cluster that was only distantly related to the two other Saccharomyces species. These results confirmed the identity of the S. boulardii strains used in this study and reaffirmed that S. boulardii and S. cerevisiae are so closely related that they likely belong to a single species.
Evaluation of S. boulardii for antimicrobial activity in comparison with S. cerevisiae
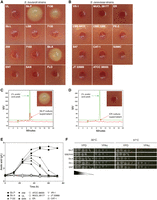
We next assessed the potential for antimicrobial activity of the S. boulardii strains in comparison with different S. cerevisiae strains by the agar-well diffusion assay using Escherichia coli MG1655 as an indicator strain. The yeast strains were propagated in yeast extract peptone dextrose (YPD) medium with 2% glucose at 37°C for 48 h, and cell-free culture supernatant was used for the assay. Out of 12 S. boulardii and 11 S. cerevisiae cell-free culture supernatants tested, those obtained from S. boulardii strains Sb.P and Sb.A produced a clear inhibition zone, while a third strain, 7136, caused mild inhibition (Fig. 1A). Except for these three strains, no clear inhibition was observed under these culture conditions with 2% glucose for any of the other S. boulardii as well as S. cerevisiae strains initially used in this study (Fig. 1A,B).
S. boulardii produces acetic acid at levels with antibacterial potency. (A,B) Agar-well diffusion assays assessing antibacterial activity of cell-free YPD with 2% glucose culture supernatants from S. boulardii strains (left panel) and S. cerevisiae strains (right panel), as visualized with plates containing E. coli MG1655 as indicator strain and staining of cells with iodonitrotetrazolium chloride (INT). (C,D) HPLC chromatograms of cell-free culture supernatants (red line) from Sb.P (S. boulardii) and ER (S. cerevisiae) compared with the 2% acetic acid standard (green line). Insets: antibacterial activity agar-well diffusion assay using E. coli MG1655 as indicator strain. (E) Acetic acid accumulation as a function of time for wild-type S. boulardii (Sb.P, Sb.A, 7136, 259, UL, SAN) (filled symbols) and S. cerevisiae strains (ATCC 38555, MUCL 28177, ER, VR-1, JT22689, CAT-1) (open symbols). (F) Spot growth assays on solid YPD (2%) and YPAc (1%, pH 5) agar medium for S. boulardii strains capable of accumulating very high acetic acid levels (Sb.P and Sb.A) versus strains with lower and transient acetic acid accumulation (Sb.L and ENT). SBERH6 is a hybrid haploid segregant derived from Sb.P and ER with very high acetic acid accumulation capacity. Gradient dark color on wedge indicates cell culture dilution strength prior to spotting.
Identification of the antimicrobial agent secreted by S. boulardii
To identify the biochemical nature of the antimicrobial action of S. boulardii, the cell-free culture supernatant from the Sb.P strain was further analyzed. This led to identification of acetic acid as the causative agent (see Supplemental Material), amounting to a concentration of 6 g/L acetic acid in the Sb.P culture supernatant as determined by HPLC (Fig. 1C). Acetic acid was virtually absent in the supernatant from S. cerevisiae Ethanol Red (ER) (Fig. 1D). The antibacterial effect on E. coli could be mimicked by adding acetic acid (6 g/L, pH 4.2) (Supplemental Fig. S2). The culture supernatant of S288c, as observed previously (Fig. 1B), yielded no zone of inhibition (Supplemental Fig. S2).
Detailed assessment of acetic acid secretion in other S. boulardii strains
Next, we performed time-course measurements of acetic acid production during aerobic growth at 37°C in cultures of all S. boulardii and some S. cerevisiae strains, with 12-h intervals over 72 h. All S. boulardii strains produced acetic acid to some extent, while it was negligible for S. cerevisiae strains (Fig. 1E). An increase in acetic acid occurred during the first 24–36 h of growth, with similar titers of 2.9 ± 0.25 g/L after 24 h. Subsequently, the Sb.P and Sb.A strains continued to accumulate acetic acid up to about 48 h, reaching a maximum of 5.30 g/L for Sb.P and 5.10 g/L for Sb.A, which remained constant up to 72 h. The other S. boulardii strains started to consume acetic acid, resulting in a lower and transient accumulation profile (Fig. 1E). Apparently, Sb.P and Sb.A are unable to consume again the acetic acid produced, explaining their much higher acetic acid production and much higher activity in the antibacterial assay, with Sb.P being the most potent for both properties.
Assessment of selected S. boulardii strains for growth capacity on acetate
Since Sb.P and Sb.A appeared unable to consume acetic acid, we assessed growth capacity on YP-acetate medium by spot assays of S. boulardii Sb.P, Sb.A, ENT, and Sb.L. All S. boulardii strains grew on acetate at 30°C, but Sb.P and Sb.A were unable to grow on acetate at 37°C (Fig. 1F). This showed that the very high acetic acid accumulation capacity of these two strains is a temperature-dependent property only manifested at the human body temperature.
Effect of acetic acid accumulation on S. boulardii cell proliferation and cell viability
Next, we have analyzed the effect of acetic acid accumulation on S. boulardii cell proliferation and viability. Three S. boulardii strains (Sb.P, Sb.L, and Enterol) were propagated alongside one S. cerevisiae strain (ER) at 37°C, and samples of the cultures were withdrawn at 12-h intervals. Acetic acid concentration, pH of the culture medium, biomass increment (measured by OD600), and cell viability were determined at each time-point (Supplemental Fig. S3). The levels of acetic acid produced by these strains confirmed the previous results. Also, the accumulation of acetic acid by the Sb.P strain closely correlated with the quasi-linear decrease in medium pH over 72 h from about 6 to 4.2. In contrast, medium pH for the remaining strains declined over 72 h only by about 1 pH unit, which is normally observed for yeast cultures grown into stationary phase. Biomass and cell viability measurements indicated that the high acetic acid accumulation of Sb.P inhibited its own proliferation and accelerated cell death. When 2 g/L acetic acid had been accumulated, Sb.P remained metabolically active since it continued acetic acid production but did not proliferate further. Cell death was observed at an acetic acid concentration of 5–6 g/L (Supplemental Fig. S3A,D), which resulted for Sb.P in an unusually low final cell viability of only 20%. On the other hand, the strains Sb.L, Enterol, and ER displayed typical growth curves for yeast, entering stationary phase after 36 h (Supplemental Fig. S3C) and maintaining near 100% cell viability (Supplemental Fig. S3D).
Obtaining a mating-competent, haploid S. boulardii derivative with high acetic acid production
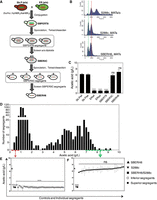
Because it showed the strongest acetic acid production and antibacterial activity, the S. boulardii Sb.P strain was selected for dissecting the genetic basis of this putative probiotic trait. To circumvent the inability of S. boulardii to sporulate, we generated an Sb.P MATa/a strain and an S. cerevisiae ER MATα/α strain. First, both copies of the HO gene were deleted in S. boulardii Sb.P, which is homothallic, to avoid autodiploidization of the final segregant. We then performed plasmid-borne galactose-induced HO expression in the Sb.P hoΔ/Δ strain to obtain Sb.P MATa/a and also in the diploid S. cerevisiae ER strain to obtain ER MATα/α. The two diploid strains were crossed to obtain a tetraploid hybrid strain, SBPERT8, which proved to be sporulation-competent. Among 145 segregants of SBPERT8, we identified SBER3C, a diploid MATa/α strain with a similar very high acetic acid production at 37°C as Sb.P. SBER3C was then sporulated and its haploid segregants scored for acetic acid production. The whole breeding strategy is summarized in Figure 2A. The haploid segregant, SBERH6 (Fig. 2B), showed highest acetic acid production (7 g/L), comparable to that of Sb.P (Fig. 2C), and was also able to grow on acetate at 30°C but not at 37°C (Fig. 1F). The other segregants of SBER3C produced variable levels of acetic acid, suggesting a polygenic basis of the phenotype (illustrated in Supplemental Fig. S4).
Origin of the superior haploid parent strain SBERH6 and phenotyping of its progenitors and segregants after crossing with the inferior haploid parent strain S288c. (A) Breeding scheme used to obtain the superior haploid strain SBERH6. (B) Determination of SBERH6 ploidy by comparison with diploid and haploid S288c strains. (C) Acetic acid production level of SBERH6 and its progenitor strains. Sb.Pwt and ERwt are wild-type Sb.P and ER strains, respectively. Sb.Paa and ERαα are derivatives of Sb.P and ER with homozygous MAT loci. SBERT8 is a tetraploid hybrid derived from Sb.Paa and ERαα, while SBER3C is a diploid segregant of SBERT8. SBERH6 is a segregant from SBER3C. Bars show mean values of two independent experiments, while error bars show standard deviation. Data were analyzed by one-way ANOVA (Dunnet correction for multiple comparisons). Significant differences for acetic acid accumulation in comparison with Sb.Pwt are indicated with asterisks, (****) P < 0.0001, (ns) nonsignificant. (D) Distribution curve of acetic acid production in segregants of the SBERH6/S288c hybrid diploid strain. (E,F) Acetic acid production of segregants selected for the inferior pool (left panel), superior pool (right panel), and control strains SBERH6, S288c, and SBERH6/S288c. Values are means from two independent experiments, while error bars represent standard deviation. Data were analyzed by one-way ANOVA (Dunnet correction for multiple comparisons). Significantly different values for acetic acid accumulation in comparison with SBERH6 are indicated with asterisks, (****) P < 0.0001, (ns) nonsignificant.
Elucidation of the genetic basis of high acetic acid production using QTL mapping
The very high acetic acid-producing haploid segregant selected, SBERH6 (MATα), was used as the superior parent in a cross with an inferior parent, the prototrophic laboratory strain, S288c (MATa). The hybrid diploid (SBERH6/S288c) showed negligible acetic acid production, like S288c (Fig. 2E,F), indicating involvement of at least one essential recessive mutation. SBERH6/S288c showed good sporulation but moderate spore viability (±50%). Acetic acid production of 549 segregants of SBERH6/S288c showed a tendency toward a bimodal distribution. However, a sizeable proportion of the segregants was located in between the two extreme tails of the distribution, displaying intermediate acetic acid production (Fig. 2D). Out of 549 segregants, 32 exhibited very high acetic acid production and were included in the superior pool (Fig. 2F). The same number of segregants producing negligible acetic acid as parent S288c was included in the inferior pool (Fig. 2E).
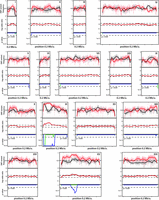
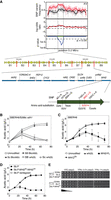
Equal quantities of cell biomass from the segregants in each pool were combined and subjected to genomic DNA extraction, obtaining 65.8 and 72.0 µg of DNA for the superior and inferior pools, respectively. The superior parent SBERH6 yielded 88.0 µg genomic DNA. Genomic DNA of both pools and superior parent was sequenced using Illumina HiSeq 2000 technology (BGI). The sequence reads were mapped to the S288c reference sequence, and variants were identified and filtered using the next-generation sequencing experience platform (NGSEP) (Duitama et al. 2014) and CLC genomic workbench (CLC Bio-Qiagen). The genomic DNA from the superior pool yielded 6,329,693 paired reads, which resulted in 97.19% overall alignment with the S288c sequence, while 6,328,957 paired reads from the genomic DNA of the inferior pool achieved 96.2% alignment. Single nucleotide variant (SNV) frequencies deviating upward from 50% in the superior pool, when compared with a hypothetical SNP frequency of 50% representing random segregation, indicated linkage to the high acetic acid phenotype. Plotting the SNV frequency (y-axis) versus the SNP chromosomal position (x-axis) revealed two major QTLs linked to the genome of superior parent strain SBERH6: QTL1 in Chromosome XI (NC_001143.9:g. 31118…231737) and QTL2 in Chromosome XV (Fig. 3). There were no complementary QTLs deviating downward from 50% and linked to the inferior parent strain S288c upon mapping with the inferior segregant pool, again providing evidence for a polygenic phenotype (Fig. 3). There was one significant QTL in the inferior pool linked to the genome of the superior parent and located at the left side of QTL1, but because of its location close to the telomere, reliability was uncertain. We concentrated further analysis on identification of the causative gene(s) within QTL1 and QTL2.
QTL mapping for very high acetic acid accumulation capacity with SNPs as genetic markers. Dot plots of SNP variant frequency from the superior pool (red) and the inferior pool (black) versus chromosomal position. Red and black lines on the dot plots represent smoothed data from the superior and inferior pools, respectively. The red line in the middle graph indicates deviation from the confidence interval. P-values (blue line) ≤ 0.05 for the difference between the smoothed lines of superior (blue line) or inferior (green line) pools at a particular locus versus 50% random segregation indicate statistically significant linkage to the genome of the superior (SBERH6) or inferior (S288c) parent at that locus. Two major QTLs (QTL1 and QTL2) with strong linkage to the genome of the superior parent are present in the first half of Chromosome XI and the first half of Chromosome XV, respectively.
Analysis of QTL1 by bulk reciprocal hemizygosity analysis (bRHA)
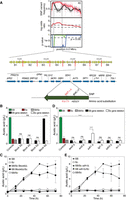
QTL1 had a length of 200,619 bp. We divided QTL1 into eight gene blocks for bulk reciprocal hemizygosity analysis (bRHA) (Fig. 4A). Each block was deleted in a reciprocal manner in Chromosome XI of the SBERH6/S288c diploid. Allele-specific PCR was used to determine whether a block of superior or inferior parent genes was deleted. Comparison of acetic acid production in the reciprocally deleted strains revealed that block 6 harbored causative genetic element(s) (Fig. 4B). This block was 25,573 bp in size and flanked by the chromosomal positions NC_001143.9:g. 156173 and 181746. The bRHA strain with block 6 from the superior parent (SBERH6/S288c block6Δ) exhibited high acetic acid production (5.65 ± 0.18 g/L), similar to SBERH6 (Fig. 4B). In contrast, the bRHA strain with block 6 from the inferior parent (SBERH6 block6Δ/S288c) produced negligible acetic acid (0.67 ± 0.06 g/L). Results for the three other blocks were negative (Fig. 4B). However, time-course determination of acetic acid production in the two bRHA strains of SBERH6/S288c revealed that block 6 only partially accounted for this phenotype. The SBERH6/S288c block6Δ strain showed a similar trend for acetic acid production as the superior parent SBERH6 up to 36 h of incubation but started to decline slightly afterward, while acetic acid production of SBERH6 continued to increase substantially (Fig. 4C).
RHA for identification of the causative gene in QTL1. (A) Overview of QTL1 dissection to nucleotide level resolution. Division of QTL1 in eight gene blocks for bRHA and genes present in block 6. Scheme of the SDH1 allele of SBERH6, depicting the two nonsynonymous SNPs in the coding region of SDH1 and their resulting amino acid substitutions in Sdh1. The causative mutation is shown in red. (B) Representative example of bRHA of acetic acid secretion with pairs of SBERH6/S288c (SB/Sc) diploid strains for the blocks 3, 4, 5, and 6 (B3–B6), indicating that block 6 contains one or more causative genes. (C) Acetic acid production as a function of time by the RHA strains with deletion of block 6, SBERH6/S288c B6Δ (•) and SBERH6 B6Δ/S288c (▪) compared to the control strains SB (SBERH6) (♦), Sc (S288c) (○), and SBERH6/S288c (SB/Sc) (□). (D) RHA with the individual genes present in block 6, identifying SDH1 as causative gene. (E) Acetic acid production as a function of time by the RHA strains for the SDH1 gene, SB/Sc sdh1Δ (SBERH6/S288c sdh1Δ) (▴) and SB sdh1Δ/Sc (SBERH6 sdh1Δ/S288c) (▾), compared to the control strains Sb (SBERH6) (♦), Sc (S288c) (○), and SBERH6/ S288c (□). Results are the means of three biological replicates for each time-point. Error bars show standard deviation at each time-point. Data were analyzed by two-way ANOVA (Tukey's multiple comparisons test). Significant differences in acetic acid accumulation between strains are indicated with asterisks, (****) P < 0.0001, (ns) nonsignificant.
Identification of the causative allele within QTL1 on Chromosome XI
Sixteen open reading frames were present in block 6 (Fig. 4A), of which APE2, SDH1, AVT3, LTV1, SDH3, and TGL1 harbored at least one missense mutation or, in the case of APE2, a frameshift mutation. These genes were prioritized as candidate genes for RHA. Pairs of hemizygous diploid SBERH6/S288c strains for each gene were constructed and tested for acetic acid production. The SBERH6/S288c sdh1Δ strain displayed a much higher acetic acid production (yielding 2.28 ± 0.75 g/L) than SBERH6 sdh1Δ/S288c, which only produced 0.66 ± 0.05 g/L (Fig. 4D). The hemizygous strains for the other five genes did not show any significant difference in acetic acid production (Fig. 4D). A time-course experiment for acetic acid production showed that SBERH6/S288c sdh1Δ behaved similarly to SBERH6/S288c block6Δ, consistent with SDH1 being the only causative allele in block 6 (Fig. 4C,E). The maximum acetic acid production level of 6.15 ± 1.04 g/L reached after 36 h by SBERH6/S288c sdh1Δ was similar to that of the superior parent strain SBERH6 (Fig. 4E), while the SBERH6 sdh1Δ/S288c strain displayed a similar, very low level of acetic acid production (0.84 ± 0.02 g/L) (Fig. 4E) as observed for the SBERH6 block6Δ/S288c strain (Fig. 4C).
Sequence analysis of SDH1 for identification of candidate causative nucleotide polymorphism(s)
Two point mutations were found in the open reading frame of SDH1 from SBERH6, which is derived from the S. boulardii Sb.P strain, being c.[604C > T]; [950T > A] and resulting in two amino acid substitutions: p.[H202Y];[F317Y]. Hence, we have named this allele sdh1H202Y,F317Y. Subsequent sequence analysis revealed that the same point mutations and amino acid substitutions were also present in SDH1 in the other 11 strains of S. boulardii (Fig. 5A). To investigate whether these SDH1 mutations were also present in S. cerevisiae strains and in other S. boulardii strains, we have screened all published genome sequences of the two yeasts for their presence. Besides the 12 S. boulardii strains from this study, publicly available whole-genome sequencing data from 992 unique strains (Strope et al. 2015; Peter et al. 2018) were mapped against the S288c reference sequence. This included 24 strains with high similarity to S. boulardii, clustered together in “subclade 3” by Peter et al. (2018). Positions with <85% of the reads indicating a single base were considered heterozygous and were assigned a degenerate base call in the reference mapped assemblies. Figure 5B shows the alignment for 18 of the S. cerevisiae strains as an example. This analysis showed that c.604C > T in SDH1, changing histidine202 to tyrosine202, is also present in some S. cerevisiae strains. However, c.950T > A, changing phenylalanine317 to tyrosine317 in Sdh1, was present in all 12 S. boulardii strains and in all 24 strains categorized as belonging to the S. boulardii subclade by Peter et al. (2018). Of the 968 additional S. cerevisiae strains analyzed, this mutation was identified only in the strain most closely related to the S. boulardii subclade, ARL (Supplemental Fig. S10). Figure 5C shows the alignment for two of the additional S. boulardii strains from Khatri et al. (2017) as an example.
Sequence comparison of selected regions in Sdh1 from S. boulardii and S. cerevisiae and identification of the causative mutation. (A) Alignment of the Sdh1 amino acid sequence in regions 181–206 and 313–321 of the S. boulardii strains and of S. cerevisiae S288c. (B) Alignment of the corresponding Sdh1 sequence of S. boulardii SBERH6 and 18 additional S. cerevisiae strains. (C) Alignment of the corresponding Sdh1 sequence of two additional S. boulardii strains. The two positions, 202 and 317, with amino acid substitutions, are indicated with black arrows while substitute amino acid residues are in red. (D) Accumulation of acetic acid as a function of time by SBERH6 (•) and by SBERH6 strains with sdh1 replaced by SDH1Y202H,Y317F (□), SDH1Y317F (▴), or sdh1Y202H (◊). (E) Accumulation of acetic acid as a function of time by Sb.P (•) and by Sb.P strains with the two copies of sdh1 replaced by SDH1Y202H,Y317F (▿) or sdh1Δ (□). Results are the means of three biological replicates for each time-point. Error bars show standard deviation at each time-point.
Functional confirmation of the sdh1H202Y,F317Y causative nucleotide(s) in SBERH6 and Sb.P
To confirm SDH1 as causative for high acetic acid production, allele exchange was performed in SBERH6 and S288c. When sdh1H202Y,F317Y in SBERH6 was replaced by its counterpart from S288c, resulting in SBERH6 SDH1Y202H,Y317F, the high acetic acid phenotype was abolished (Fig. 5D), while it was restored by reintegration of the original sdh1H202Y,F317Y (Supplemental Fig. S5). To identify the causative SNP within SDH1, hybrid alleles, sdh1Y202H (with only 604T>C) and SDH1Y317F (with only 950A>T), were introduced in SBERH6. SBERH6 SDH1Y317F lacked acetic acid production, similar to SBERH6 SDH1Y202H,Y317F, while SBERH6 sdh1Y202H showed similar accumulation as SBERH6 (Fig. 5D). This indicated that the c. 950T>A mutation, which results in the change of phenylalanine317 to tyrosine317, is the only causative SNP in sdh1 of SBERH6. Conspicuously, this is the unique sdh1 SNP found in all S. boulardii strains and in none of the surveyed S. cerevisiae strains.
Given that SBERH6 was derived from a tetraploid hybrid between Sb.P and ER (Fig. 2A), SBERH6 likely harbored a mosaic genome, with regions from Sb.P and from ER. To confirm the causative character of the superior SDH1 allele also in the original Sb.P strain, its two SDH1 copies were replaced by the inferior allele from S288c. This eliminated high acetic acid production (Fig. 5E), and it was re-established by reintegration of the original superior SDH1 allele (Supplemental Fig. S5). This effect was comparable to that observed upon allele exchange in SBERH6 (Fig. 5D). Deletion of SDH1 in Sb.P did not affect high acetic acid accumulation (Fig. 5E), suggesting that its superior allele sdh1H202Y,F317Y may be a loss-of-function allele.
Identification of the causative allele in QTL2 on Chromosome XV
To identify in QTL2 on Chromosome XV the causative allele for high acetic acid accumulation, we performed bRHA with a new hybrid strain that was obtained by crossing SBERH6 with S288c sdh1H202Y,F317Y. The resulting SBERH6/S288c sdh1H202Y,F317Y strain has two copies of the superior sdh1H202Y,F317Y allele, which is required for high acetic acid accumulation because the allele is recessive. To narrow down the region with the highest linkage in QTL2, we first performed fine-mapping using allele-specific PCR. The variant frequency and corresponding P-value were calculated after scoring selected SNP markers in individual segregants from both pools (Supplemental Fig. S6). This confirmed strong linkage in the center of QTL2 to the genome of superior parent SBERH6. To perform bRHA, the region from chromosomal position NC_001147.9:g. 278057 to NC_001147.9:g. 433375 was divided into nine blocks, after which each block was deleted in a reciprocal manner in Chromosome XV of SBERH6/S288c sdh1H202Y,F317Y (Fig. 6A). Allele-specific PCR was used to determine whether the deleted block was from the superior or inferior parent, and the strains with reciprocal deletion were compared for acetic acid accumulation. Only for block 8, a clear difference between the two hemizygous strains was observed (Fig. 6B). SBERH6 block8Δ/S288c sdh1H202Y,F317Y showed low, transient acetic acid accumulation comparable to that of SBERH6/S288c sdh1H202Y,F317Y, while SBERH6/S288c sdh1H202Y,F317Y block8Δ showed high, permanent acetic acid accumulation comparable to that of SBERH6 (Fig. 6B). Block 8 (NC_001147.9:g. 394837 to NC_001147.9:g. 433375) contained 10 genes: AKR2, YOR034C-A, SHE4, PEP12, CYC2, HIR2, CKB2, GLO4, CUE5, and WHI2. Since WHI2 contained several nonsynonymous SNPs, is known to regulate STRE (STress Response Element)-mediated gene expression (Kaida et al. 2002), and was previously implicated in acetic acid tolerance (Chen et al. 2016), it was investigated individually by RHA, while the remaining genes were combined in one block, block 8.1. Acetic acid accumulation in the two RHA strains for WHI2 showed a comparable difference to that observed for the whole block 8, consistent with the superior allele being derived from superior parent SBERH6 and the inferior allele from inferior parent S288c sdh1H202Y,F317Y (Fig. 6B). For the RHA strains of block 8.1, no difference in acetic acid accumulation was observed (Supplemental Fig. S7).
RHA for identification of the causative gene in QTL2. (A) Overview of QTL2 dissection to nucleotide level resolution. Division of QTL2 into nine gene blocks for bRHA and genes present in block 8. (B) Acetic acid accumulation by the bRHA strains for block 8 and for WHI2. The bRHA strains for block 8 or WHI2 are shown as SB Block8Δ or SB whi2Δ and Sc Block8Δ or Sc whi2Δ for reciprocal deletions in the SBERH6 and S288c genome, respectively, in the SBERH6/S288c sdh1H202Y,F317Y background. Strains: SBERH6/S288c sdh1H202Y,F317Y (□), SBERH6/S288c sdh1H202Y,F317Y B8Δ (▴), SBERH6 B8Δ/S288c sdh1H202Y,F317Y (▾), SBERH6 whi2Δ/S288c sdh1H202Y,F317Y (▿), and SBERH6/S288c sdh1H202Y,F317Y whi2Δ (▵). (C) Acetic acid accumulation by SBERH6 strains with modifications of WHI2. Strains: SBERH6 (○), SBERH6 whi2Δ (▴), SBERH6 WHI2-FL (WHI2*287S) (▪), and SBERH6 WHI2Sc (S288c allele of WHI2) (▾). (D) Acetic acid accumulation by Sb.P with its two copies of whi2S287* replaced by two copies of WHI2Sc. Strains: Sb.P (•), Sb.P WHI2Sc/WHI2Sc (▪), and Sb.P whi2287* reintegrant (○). Results are the means of three biological replicates for each time-point. Error bars show standard deviation at each time-point. (E) Spot growth assays on solid YPD (2%), YPAc (0.5%, pH 5), and YPAc (1%, pH 5) agar medium at 30°C and 37°C for S288c, Sb.P, and Sb.P WHI2Sc/WHI2Sc. Gradient dark color on wedge indicates cell culture dilution strength prior to spotting.
Identification of a unique S. boulardii SNP in the superior WHI2 allele
The open reading frame of WHI2 from strain SBERH6 contained 12 SNPs, of which six were nonsynonymous (Table 1). Eleven of these SNPs also occur in WHI2 in some of the 992 S. cerevisiae strains (Strope et al. 2015; Peter et al. 2018) surveyed. One single SNP, the nonsense mutation c.860C > G, which generates a premature stop codon (S287*), did not occur in any of the S. cerevisiae strains but was present in 11 of the 12 S. boulardii strains used in our study (all except strain LSB; see also below in the section on “Heterozygosity status”) and also in all 24 strains categorized as belonging to the S. boulardii subclade by Peter et al. (2018). As an example, we show an alignment of the WHI2 sequence from 45 S. cerevisiae strains, revealing that 11 of the SNPs also occur in at least one of them (Supplemental Table S2). Sb.P and Sb.A were homozygous for this SNP (S287*), while all other S. boulardii strains were heterozygous, except for strain CIT among the isolates analyzed by Peter et al. (2018), which was homozygous (see also below in the section on “Heterozygosity status”), and one strain, LSB, lacked the SNP altogether, keeping the 860C nucleotide of S. cerevisiae (Table 1). Hence, we identified a new unique SNP that is present in nearly all S. boulardii strains and occurs in one or two copies, possibly causally linked to low or high acetic acid accumulation capacity, respectively.
Occurrence of SNPs in WHI2 for S288c, SBERH6, and 12 S. boulardii strains
Identification of the unique S. boulardii SNP as the causative SNP in the superior WHI2 allele
When the mutant WHI2 allele in SBERH6 was replaced by the wild-type WHI2 allele from S288c, WHI2Sc, a transient accumulation of acetic acid was observed with a maximum after 36 h as opposed to the continuous accumulation of acetic acid in the SBERH6 strain (Fig. 6C). Since the nonsense mutation c.860C > G in SBERH6 results in a truncated, possibly inactive protein, we also deleted WHI2 in SBERH6. The SBERH6 whi2Δ (whi2::NatMX4) strain indeed showed a very similar accumulation of acetic acid as SBERH6 (Fig. 6C). This suggested that the nonsense mutation c.860C > G in the superior WHI2 allele might be the causative mutation. To evaluate this possibility, a modified WHI2 allele, WHI2-FL (WHI2*287S), was introduced into SBERH6. This allele contained all SNPs present in the promoter, ORF, and terminator of WHI2 from SBERH6, except the nonsense mutation c.860C > G. The SBERH6 WHI2-FL strain showed transient acetic acid accumulation like SBERH6 WHI2Sc (Fig. 6C), while reintegration of the original whi2SBERH6 mutant allele restored continuous acetic acid accumulation (Supplemental Fig. S5). This indicated that the c.860C > G mutation was indeed the causative SNP in the whi2 allele of SBERH6.
Importance of the whi2S287* copy number for high acetic acid accumulation and assimilation
We confirmed that the copy number of whi2S287* was responsible for continuous accumulation of high levels of acetic acid in Sb.P, by exchanging the two copies of whi2S287* with its counterpart from S288c, generating Sb.P WHI2Sc/WHI2Sc. This caused the high acetic acid accumulation profile of Sb.P to change into the lower and transient acetic acid accumulation profile of the other S. boulardii strains, while reintegration of the original mutant whi2S287* allele restored the phenotype (Fig. 6D). The Sb.P WHI2Sc/WHI2Sc strain also grew at 37°C on YP-acetate (1%) plates (Fig. 6E). At a lower acetate concentration (0.5%), the growth defect at 37°C caused by whi2S287* was only partial (Fig. 6E).
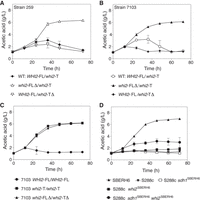
Next, we investigated whether the very high acetic acid accumulation phenotype of Sb.P could be transferred to other S. boulardii strains with lower and transient acetic acid production by modification of the WHI2 allele. For that purpose, the WHI2 gene was deleted in a reciprocal manner in S. boulardii strain 259, either eliminating the superior whi2S287* (encoding truncated Whi2) or the inferior WHI2-FL allele (encoding full-length Whi2). The strain in which only the superior whi2S287* allele was deleted, 259 WHI2-FL/whi2-TΔ, maintained its lower and transient acetic acid accumulation, whereas the strain in which the inferior WHI2-FL allele had been deleted, 259 whi2-FLΔ/whi2-T, gained very high acetic acid accumulation (Fig. 7A). Comparable results were obtained with another S. boulardii strain with lower and transient acetic acid production, 7103 (Fig. 7B). In both strains, 259 and 7103, deletion of the whi2S287* allele caused a further reduction in acetic acid production, indicating that the truncated whi2S287* gene product may retain some activity.
Confirmation of the functional relevance of WHI2 for the level of acetic acid production in other S. boulardii strains. (A) Acetic acid production in strain 259 with a single deletion of the full-length WHI2 allele (whi2-FLΔ/whi2-T) or the mutant whi2S287* allele (WHI2-FL/whi2-TΔ). Strains: wild-type 259 WHI2-FL/whi2-T (♦), 259 whi2-FLΔ/whi2-T (▵), and 259 WHI2-FL/whi2-TΔ (▿). (B) Acetic acid production in strain 7103 with a single deletion of the full-length WHI2 allele (whi2-FLΔ/whi2-T) or the mutant whi2S287* allele (WHI2-FL/whi2-TΔ). Strains: wild-type 7103 WHI2-FL/whi2-T (○), 7103 whi2-FLΔ/whi2-T (▴), and 7103 WHI2-FL/whi2-TΔ (♦). (C) Acetic acid production in strain 7103 with two full-length WHI2 alleles (WHI2-FL), two mutant whi2S287* alleles (whi2-T), or a double deletion of both alleles. Strains: 7103 WHI2-FL/WHI2-FL (•), 7103 whi2-T/whi2-T (▪), and 7103 whi2-FLΔ/whi2-TΔ (▴). (D) Acetic acid production in S. cerevisiae S288c containing the causative alleles sdh1H202Y,F317Y (sdh1SBERH6) and/or whi2S287* (whi2SBERH6) from S. boulardii. Strains: SBERH6 (▴), S288c (▾), S288c sdh1SBERH6 (□), S288c whi2SBERH6 (▪), and S288c sdh1SBERH6 whi2SBERH6 (•). Results are the means of three biological replicates for each time-point. Error bars show standard deviation at each time-point.
We also constructed strains that were homozygous for the WHI2-FL allele encoding full-length Whi2, 7103 WHI2-FL/WHI2-FL, or for the superior whi2S287* allele encoding truncated Whi2, 7103 whi2-T/whi2-T. Strain 7103 whi2-T/whi2-T showed the same very high acetic acid accumulation as Sb.P. On the other hand, strain 7103 WHI2-FL/WHI2-FL maintained the lower and transient acetic acid accumulation of strain 7103 (Fig. 7C). These results show that exchange of the superior whi2S287* allele for the inferior WHI2-FL allele in an S. boulardii strain with lower and transient acetic acid production, resulting in a strain with two whi2S287* alleles, is enough to convert it into a strain with very high acetic acid accumulation.
Transfer of the high acetic acid accumulation capacity to S. cerevisiae S288c by introduction of the SDH1 and WHI2 superior alleles
To determine whether S. boulardii sdh1H202Y,F317Y and whi2S287* were sufficient to transfer, at least to some extent, the high acetic acid accumulation phenotype to S. cerevisiae, we integrated the two alleles separately as well as together in S288c. While separate introduction did not affect very low acetic acid production in S288c, combined introduction resulted in acetic acid accumulation up to nearly half (2.98 ± 0.8) of the level in SBERH6 (6.98 ± 0.07) (Fig. 7D). The absence of full establishment of very high acetic acid production may indicate that the S. boulardii Sb.P and Sb.A strains still contain one or more additional mutations required for this phenotype or that the laboratory strain S288c contains compromising mutations not present in Sb.P and Sb.A. In spite of this, the result further supports that very high acetic acid production in S. boulardii Sb.P and Sb.A is a polygenic trait, requiring the simultaneous presence of at least two genetic polymorphisms.
Higher sugar levels lead to very high acetic acid accumulation in all S. boulardii strains but not in most S. cerevisiae strains
To screen for antibacterial activity in a larger set of S. cerevisiae strains, we first optimized the assay so that we could detect antibacterial activity in the S. boulardii strains with lower, transient acetic acid accumulation. Increasing the glucose level from 2% to 4% resulted in similarly high antibacterial activity (Supplemental Fig. S8A) and acetic acid accumulation (Supplemental Fig. S8C vs. S8D) in all S. boulardii strains. Such high antibacterial activity (Supplemental Fig. S8B) and acetic acid accumulation (Supplemental Fig. S8C–E) were not observed in the 304 S. cerevisiae strains tested for both properties, although a minority of S. cerevisiae strains also produced intermediate and some even higher acetic acid levels approaching those produced by the S. boulardii strains (Supplemental Fig. S8E). As the only exception, one mutant haploid S. cerevisiae lab strain, derived from M5, produced an equal level of acetic acid as S. boulardii.
Phylogenetic analysis based on whole-genome sequence analysis of all S. boulardii strains in comparison with whole-genome–sequenced S. cerevisiae strains
We have made a neighbor-joining tree based on whole-genome SNP analysis after mapping of Illumina whole-genome sequencing data, using BioNumerics 7.6 (Applied Maths) of 352 yeast strains belonging to Clade 1.Wine/European, as well as 12 S. boulardii strains, according to Peter et al. (2018) (Supplemental Fig. S9). The 12 S. boulardii strains clearly cluster together with the Subclade 3 strains, including known S. boulardii strains. Strain ARL is the S. cerevisiae strain most closely related to all S. boulardii strains.
Heterozygosity status of the WHI2 chromosomal region in all S. boulardii strains
We have made an analysis of the heterozygosity status of 28 SNP positions in a region of Chromosome XV at the WHI2 locus (Supplemental Fig. S10). Strains Sb.P and Sb.A have loss of heterozygosity in an extended region resulting in homozygosity for the whi2S287* allele, whereas strain CIT is only homozygous for the whi2S287* allele and not for the rest of the region. Strains LSB and ARL have loss of heterozygosity at this position resulting in homozygosity for the WHI2 wild-type allele. The Enterol strain is representative for all 31 other S. boulardii strains analyzed, showing heterozygosity in the entire region.
Discussion
S. boulardii is a worldwide commercialized probiotic yeast with well-established efficacy in clinical trials (Kotowska et al. 2005; McFarland 2006, 2010; Villar-García et al. 2015). It has remained unclear why and how the yeast S. boulardii would exert a specific probiotic effect as opposed to S. cerevisiae, which is so closely related in genome sequence that the two are now considered a single species. On the other hand, there are apparently no solid comparative studies on the probiotic potency of S. cerevisiae versus S. boulardii in clinical trials. Hence, it has remained unclear whether S. boulardii might possess unique traits compared to S. cerevisiae that are important for its presumed superior probiotic potency.
There have been cases of commercial probiotic preparations lacking purity or containing different organisms than anticipated (Fasoli et al. 2003; Theunissen et al. 2005; Huys et al. 2006). Also, some of the strains in our collection (Sb.A, Sb.L, and Sb.P) were from unknown origin (van der Aa Kühle et al. 2005). Hence, we determined the identity of all S. boulardii strains in our collection using AFLP (MacKenzie et al. 2008) and whole-genome sequence analysis (Supplemental Table S1). This showed that the 12 S. boulardii strains were very closely related, forming a single cluster in the phylogenetic tree (Supplemental Fig. S1). They were much more closely related to all S. cerevisiae strains than to S. mikatae and S. paradoxus, providing further evidence that S. boulardii is conspecific with S. cerevisiae (van der Aa Kühle and Jespersen 2003; Fietto et al. 2004).
Antimicrobial action via antimicrobial peptide or organic acid secretion is regarded as a key probiotic property (Boirivant and Strober 2007; Vanderpool et al. 2008; Ciorba 2012). S. boulardii has been reported to secrete medium chain fatty acids, mainly capric acid, with bioactivity against Candida albicans hyphae and biofilm formation (Krasowska et al. 2009; Murzyn et al. 2010). However, to the best of our knowledge, the secretion of diffusible antimicrobial agents by S. boulardii against bacterial pathogens has never been described. All the S. boulardii strains tested secreted an antimicrobial agent with an inhibitory effect on E. coli MG1655, an enteric bacterial pathogen. We identified it as acetic acid, which explained the strong medium acidification to pH 4.2 at 37°C. Low pH is a crucial factor for short-chain organic acids to act as antimicrobial agents since the proportion of the protonated, uncharged form strongly increases at pH values below the pKa. The uncharged form easily diffuses into cells, causing aberrant intracellular acidification (Salmond et al. 1984; Lambert and Stratford 1999; Takahashi et al. 1999). The capacity of Sb.P for medium acidification by secretion of high acetic acid levels at 37°C suggests that it may compromise metabolic activity and proliferation of enteric pathogens in local microenvironments at the brush border in the gut. Although higher temperature is known to enhance acetic acid production by S. cerevisiae (Woo et al. 2014), the production of such high quantities of acetic acid (about 6 g/L) as observed in our study for S. boulardii has never been reported for S. cerevisiae. Acetic acid accumulation by Sb.P was so high that it caused inhibition of its own growth and loss of cell viability (Supplemental Fig. S3C,D). The cell-free supernatant from the other S. boulardii strains that did not show sustained production of acetic acid over time were not inhibitory to the E. coli indicator strain under the conditions with YPD and 2% glucose initially used. The same was observed for the cell-free supernatant from S. cerevisiae strains. Hence, the strong antimicrobial activity of the cell-free culture supernatant of Sb.P can be attributed to the high level of acetic acid and the low pH. Subsequent work showed that, in the presence of a higher glucose level, all S. boulardii strains produce very high levels of acetic acid resulting in very high probiotic potency. This unique property of vigorous acetic acid production may at least in part explain the effectiveness of S. boulardii as a probiotic in the gut and also its uniqueness as the only variety selected for commercial use as a probiotic among all strains now considered to belong to the species S. cerevisiae.
The secretion of acetic acid by S. boulardii in the gut may have other advantages to the host. Acetate stimulates the expansion and build-up of T regulatory cells, facilitates goblet cell proliferation and up-regulates genes related to mucus secretion, inhibits the proinflammatory cytokine CXCL8, and serves as a substrate for the production of butyrate by gut microbiota (Engevik and Versalovic 2017). A possible counterargument against its presumed benefit could be that production of acetic acid by S. boulardii is largely restricted to aerobic conditions (Supplemental Fig. S11), at least under our assay conditions. However, noninvasive measurement of oxygen tension along the gastrointestinal tract revealed a marked oxygen gradient from the proximal to the distal gastrointestinal tract (He et al. 1999), as well as the existence of a radial gradient with high O2 levels close to the epithelial surface and very low O2 levels in the center of the gut lumen (Albenberg et al. 2014). This suggests that microorganisms colonizing the intestinal epithelium at the brush border have similar oxygen availability as the cells of the epithelium. Moreover, oxygen availability levels in the gastrointestinal tract are known to increase significantly under conditions compromising the normal gut microbiota, such as antibiotic treatment (Rivera-Chávez et al. 2016, 2017; Vacca 2017) and enteropathogenic infections (Rivera-Chávez et al. 2017). This suggests that enteric disorders for which the efficacy of S. boulardii is well established, such as antibiotic-associated diarrhea (McFarland et al. 1995; Duman et al. 2005), C. difficile infections (McFarland et al. 1994), and acute adult diarrhea (McFarland 2010), may provide an appropriate aerobic environment for maximal acetic acid secretion by S. boulardii.
The time-course measurement showed that acetic acid production by the Sb.P strain was not compromised by growth limitation. Upon growth arrest at 12 h, apparently due to inhibition by the accumulated acetic acid, the acetic acid production continued unabated (Supplemental Fig. S3A,C). It only ceased at 48 h, probably due to declining cell viability (Supplemental Fig. S3A,D). Acetic acid production in the absence of growth is significant since a probiotic must be effective at its site of action (Schrezenmeir and de Vrese 2001; Hill et al. 2014) and a permissive environment for cell proliferation may not always be available in the gut.
In the initial test, only the Sb.P and Sb.A strains showed in vitro antimicrobial activity. Subsequent work, however, showed that all S. boulardii strains produce acetic acid but that the other strains are able to consume it, as opposed to Sb.P and Sb.A. As a result, Sb.P and Sb.A accumulated much more acetic acid than the other strains, which explains their much greater antimicrobial potency. Analysis of the growth capacity on acetate of the Sb.P and Sb.A strains, as well as Sb.P derivative SBERH6, revealed that they were unable to grow on acetate specifically at 37°C as opposed to the other S. boulardii strains that grew on acetate both at 30°C and 37°C. Hence, the presence of two copies of the whi2S287* allele appears to cause a temperature-sensitive acetate utilization defect resulting in the continuous, very high acetic acid accumulation, a property possibly highly relevant for probiotic potency that is only manifested at regular body temperature.
The sporulation defect of S. boulardii has been a major obstacle for any genetic analysis. In the present work, we have circumvented this problem by deleting first the two copies of the HO gene in S. boulardii to make it heterothallic, we then made it homozygous for mating type and crossed this strain with an S. cerevisiae strain made homozygous for the complementary mating type, so as to obtain a hybrid heterothallic strain. This strain was able to sporulate, enabling us to generate a mating-competent haploid hybrid strain, SBERH6, displaying the same high acetic acid accumulation capacity as Sb.P. This innovative strategy provides an efficient means by which haploid strains with a specific trait of interest can be obtained from sporulation-defective diploid or polyploid S. cerevisiae strains. It enables subsequent genetic analysis of the trait of interest on the condition that the haploid strain can be mated with an unrelated strain to obtain a sporulation-competent diploid strain. This was the case for SBERH6, so that we could then apply the polygenic analysis platform based on pooled-segregant whole-genome sequence analysis. This has now been successfully used to elucidate the polygenic basis of many traits in S. cerevisiae (Liti and Louis 2012; Swinnen et al. 2012; Hubmann et al. 2013; Pais et al. 2013; Yang et al. 2013; Meijnen et al. 2016; Trindade de Carvalho et al. 2017). The 549 segregants we obtained from the hybrid diploid SBERH6/S288c showed a distribution of acetic acid production resembling a bimodal distribution, although a significant number of segregants showed an intermediate phenotype. This distribution is consistent with a low number of causative genes involved. Subsequent work showed that the presence of two copies of sdh1H202Y,F317Y in all S. boulardii strains is essential for significant acetic acid production, much higher than that observed in S. cerevisiae, while the presence of whi2S287* in one or two copies determines the difference between transient and moderate versus continuous and high acetic acid accumulation. Hence, the interplay between the SDH1 and WHI2 alleles appears to be responsible for the overall shape of the segregant distribution curve and the divergence between different S. boulardii strains. Very high acetic acid accumulation capacity is thus a polygenic trait caused by two major genetic elements, and the slight variation observed in the segregants is likely due to an unknown number of minor genetic elements.
SDH1 encodes the flavoprotein subunit of the succinate dehydrogenase complex, which functions in the tricarboxylic acid (TCA) cycle and mitochondrial respiratory chain by coupling succinic acid oxidation to the transfer of electrons to ubiquinone (Chapman et al. 1992; Kim et al. 2012). Deletion of SDH1 or other genes encoding subunits of the succinate dehydrogenase complex is known to cause higher acetic acid production in S. cerevisiae (Romano and Kolter 2005; Szeto et al. 2010; Yoshida and Yokoyama 2012). Acetic acid was secreted by the BY4741 sdh1Δ strain at 30°C, yielding 60 mM (3.6 g/L) after 72 h (Romano and Kolter 2005; Szeto et al. 2010; Yoshida and Yokoyama 2012). In our work, Sb.P did not show any acetic acid overproduction at 30°C but yielded about 6 g/L at 37°C after 72 h. Hence, the sdh1H202Y,F317Y allele, which is essential for high acetic acid production at 37°C, could actually be a temperature-sensitive allele causing malfunctioning of the succinate dehydrogenase complex at 37°C.
The link between defective succinate dehydrogenase and high acetic acid production could arise in different ways. The Sdh defect can compromise oxidation of succinic acid to fumaric acid, causing shortage of oxaloacetic acid and thus reduced initiation of the TCA cycle. This can result in accumulation of acetyl-CoA and, through further conversion by the mitochondrial acetyl-CoA hydrolase (Buu et al. 2003), lead to production of acetic acid. Alternatively, pyruvate could accumulate because of the compromised TCA cycle, leading to higher levels of cytosolic acetaldehyde and, through conversion by aldehyde dehydrogenase, to higher levels of acetic acid.
WHI2 encodes a protein with phosphatase activator activity that forms a complex with the plasma membrane Psr1 phosphatase and is localized at the cell periphery. It is involved in multiple processes, including cell cycle regulation, cell proliferation, the general stress response, endocytosis, organization of the actin cytoskeleton, and amino acid sensing (Saul and Sudbery 1985; Radcliffe et al. 1997; Chen et al. 2018; Teng and Hardwick 2019). Whi2 is required for full activation of STRE-mediated gene expression via dephosphorylation of the transcription factor Msn2 (Kaida et al. 2002), which is important for proper degradation of misfolded proteins (Comyn et al. 2017). Overexpression of Whi2 was shown to improve acetic acid resistance, while its absence reduces it (Chen et al. 2016). Although the whi2Δ strain shows multiple other defects, a connection with acetic acid production is not straightforward. On the other hand, since the causative whi2S287* allele contains a premature stop codon, its effect may be similar to that of WHI2 deletion. Hence, the increased acetic acid sensitivity of Sb.P and Sb.A, having two copies of whi2S287*, may explain their inability to consume the high acetic acid levels accumulated as opposed to the other S. boulardii strains with only one whi2S287* allele. A crucial observation in this respect was that the strains with two copies of whi2S287* were able to grow on lower levels of acetic acid at 37°C (Fig. 6E), showing that they were not deficient in acetic acid assimilation per se. They were only deficient in consuming high levels of acetic acid at 37°C, possibly due to their reduced acetic acid tolerance. The whi2S287* allele may therefore act indirectly through acetic acid inhibition of its consumption, resulting in a higher final level of acetic acid accumulation. This explanation would also imply that none of the two causative mutations has to be a temperature-sensitive mutation. Instead, the yeast may simply be more sensitive to acetic acid at high temperature because of the whi2S287* mutation. The higher acetic acid sensitivity of Sb.P and Sb.A may also contribute to their early growth arrest, allowing more carbon to be channelled into acetic acid production instead of biomass formation. Secondary missense mutations have been reported in the WHI2 locus of strains from the yeast deletion collection (Mendl et al. 2011; van Leeuwen et al. 2016; Comyn et al. 2017). Deletion of STE20, NUC1, APM2, OTU2, GCN20, SET2, URA1, and SGN1 is associated with an elevated frequency of nonsense mutations in WHI2 (Teng et al. 2013). We could not find any obvious deleterious or nonsense SNPs in these genes in S. boulardii Sb.P, but this does not exclude that a missense SNP within one of these genes has driven the emergence of the c.860C > G SNP in WHI2.
Identification of the causative SNPs in SDH1 and WHI2 revealed one SNP in each, c.[950T > A] translating into p.[F317Y] and c.860C > G translating into p.S287Ter, respectively, that were unique for S. boulardii and absent in virtually all sequenced S. cerevisiae strains. Hence, our work has identified the first genetic signature for S. boulardii, which is so closely related in genome sequence to S. cerevisiae that the two are considered to belong to the same species. This unique genetic signature provides for the first time an easy way to identify new S. boulardii strains isolated from nature. All S. boulardii strains currently available are probably derived from the original strain(s) isolated by Boulard. Moreover, the unique polymorphisms in S. boulardii are responsible for a conspicuous physiological difference—high acetic acid production—that, because of its antimicrobial potency, may have conferred an advantage to S. boulardii in certain aerobic ecological niches. Because of its much higher toxicity compared to ethanol, acetic acid production would be advantageous in environments with lower sugar concentrations in which only low, nontoxic ethanol levels can be accumulated. It may also have been the reason why S. boulardii has been selected and successfully employed worldwide as a probiotic. If the acetic acid production capacity is important for its probiotic potency, our work opens up potential for selection, breeding, or engineering of S. boulardii strains with further improved probiotic potency or even for transfer of this trait of antimicrobial potency to S. cerevisiae strains.
Introduction of the superior sdh1H202Y,F317Y and whi2S287* alleles in S288c, however, only resulted in about 50% of the very high acetic acid accumulation observed in Sb.P and Sb.A (Fig. 7D). This can be attributed to the polygenic nature of the trait and thus to the requirement of additional causative genes present in minor QTLs. One possibility is that minor QTLs harbor redundant causative genes, of which one is required to function cooperatively with the causative SDH1 allele from Sb.P. Another possibility is that one of the major QTLs, QTL1 or QTL2, harbors another minor causative allele. Combinations of these two possibilities are also possible. Since laboratory yeast strains like S288c harbor multiple debilitating mutations, which are absent in natural and industrial S. cerevisiae strains, one of these might compromise maximal acetic acid production capacity and therefore needs to be complemented by the corresponding wild-type gene from S. boulardii.
In conclusion, we have discovered a novel, specific trait of the yeast S. boulardii, high acetic acid production, that may be of high relevance for its probiotic potency. We have elucidated the polygenic basis of this property and explained the quantitative difference in acetic acid production between S. boulardii strains. This novel trait is due to two specific point mutations which are unique to S. boulardii and provide the first specific genetic signature for S. boulardii versus S. cerevisiae. Our work provides a possible explanation for the selection of S. boulardii as the only probiotic yeast as opposed to the very closely related, likely conspecific S. cerevisiae strains.
Methods
The strains used in this work are listed in Supplemental Table S1. Media and culture conditions, general molecular biology methods, random spore isolation, and DNA preparation for amplified fragment length polymorphisms (AFLPs) determination are detailed in Supplemental Methods.
Yeast culture conditions for antimicrobial and acetic acid production assays
Overnight yeast precultures were adjusted to an OD600 of 0.2 in 50 mL YPD (with 2% glucose) in a 300-mL Erlenmeyer flask. Flasks were incubated by shaking at 200 rpm and 37°C in a shaking incubator for 48 h. To obtain cell-free culture supernatants, aliquots of yeast cultures were withdrawn from the flasks and centrifuged at maximum speed (14,000 rpm) for 5 min. The supernatants were used for agar-well diffusion assays or subjected to HPLC for determination of acetic acid concentrations. For time-course measurements, samples were withdrawn from the cultures every 12 h for further analysis.
Agar-well diffusion assays
For the agar-well diffusion assays, 25 mL molten soft Mueller Hinton agar (7.5 g bacto agar/L Mueller Hinton broth) were inoculated with 5 × 104 cells/mL E. coli indicator strain. This was followed by addition of the bacterial growth indicator, iodonitrotetrazolium chloride (in 50% methanol), to a final concentration of 0.2 mg/mL and brief vortexing. A square petri dish containing 80 mL solidified Muller Hinton agar was then overlaid with the molten top agar. The top agar was allowed to solidify, after which nine wells (in 3 × 3 format) were punched into both agar layers using a 12-mm sterile cork borer. The resulting agar discs were carefully removed from each well with a pair of sterile tongs and discarded. Each well was then filled with ∼700 µL of yeast culture supernatant. All agar-well plates were incubated at 37°C for 12–18 h.
Growth assays by spot dilution series for assessment of acetate utilization capacity
The strains were propagated in 3 mL YPD broth at 30°C for 1 d. A dilution of each culture was prepared in sterile Milli-Q water to an OD600 of 0.5. A 5-µL 10× dilution series (100-10−5) of each strain was spotted on YPD as well as on yeast extract peptone +1% potassium acetate (YPAc; pH 5). Both YPD and YPAc plates were incubated at 30°C and 37°C.
HPLC determination of acetic acid, yeast ploidy determination by flow cytometry, and yeast viability determination were performed as detailed in Supplemental Methods.
Deletion of the HO endonuclease gene and mating type switch
The two copies of the HO endonuclease gene in Sb.P were deleted and the heterothallic Sb.P and ER strains mating type switched to strains homozygous for mating type as detailed in Supplemental Methods.
DNA isolation, Illumina sequencing, SNP calling, and QTL mapping
High molecular weight DNA was isolated for sequencing as detailed in Supplemental Methods. The isolated DNA was submitted to Illumina HiSeq 2000 technology (BGI) with libraries of 500 bp and paired-end reads of 101 bp. The short-read sequences were mapped against the S288c reference sequence, and all variants (SNPs and small indels) were identified and quality filtered using NGSEP (Duitama et al. 2014). In parallel, CLC genomic workbench (CLC Bio-Qiagen) was used to map the reads in order to allow easy comparison of read mappings to the annotated genome of S288c.
Bulk reciprocal hemizygosity analysis of QTL1
QTL1 (NC_001143.9: g.31118…231737) was defined as the segment of DNA on Chromosome XI where the difference between the average SNV of the superior pool and the average of 50% for random segregation assumed statistical significance (P-value ≤ 0.05). QTL1 was split into eight blocks of genes (∼25 kbp for each block). Blocks 3, 4, 5, and 6 were deleted separately in a reciprocal manner in the hybrid diploid (SBERH6/S288c). Each deletion was achieved using a split geneticin resistance marker (KanMX4) knockout cassette. The cassette was constructed by adaptor-mediated fusion of PCR-amplified left and right flanking sequences (between 400 and 700 bp) for each block, with the left and right fragments of a KanMX4 marker, respectively. The hybrid diploid (SBERH6/S288c) was subsequently transformed with the two fragments of the KanMX4 marker specifically constructed for each block.
Reciprocal hemizygosity analysis of individual genes in QTL1
For RHA of the individual genes in QTL1, the exact ORF of the left and right flanking sequences and the prioritized genes in QTL1 (APE2, SDH1, AVT3, LTV1, SDH3, and TGL1) were deleted with KanMX4. The methodology used for cassette construction and transformation of the strains is the same as described above for bRHA. The nonessential genes APE2, SDH1, AVT3, LTV1, and TGL1 were deleted in the haploid backgrounds of SBERH6 and S288c. Successful transformants were assessed for correct integration of the cassette at each locus by PCR. S288c transformants with the right integration at each locus were subsequently crossed with SBERH6 and vice versa to obtain reciprocal hemizygous strains for each gene. The only essential gene, SDH3, was deleted in the hybrid diploid (SBERH6/S288c) background. Successful transformants harboring an SDH3 deletion in one parental chromosome were genotyped using allele-specific PCR.
Reciprocal hemizygosity analysis of QTL2
For RHA of QTL2, the entire block (bRHA) or the exact ORF was deleted with NatMX4 in the diploid SBERH6/S288c sdh1H202Y,F317Y hybrid strains. The marker was amplified from plasmid pTOPO-G1-NatMX4-G1 with primers containing 50-bp tails, homologous to the regions flanking the targeted locus, as described by Baudin et al. (1993). Correct integration of the marker was confirmed by PCR and the remaining, nondeleted allele was identified by allele-specific PCR.
Genotyping by allele-specific PCR and site-directed genetic modification using the CRISPR/Cas9 technology were performed as described in Supplemental Methods.
Data access
All sequence data have been submitted to NCBI's BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA542592.
Competing interest statement
VIB and KU Leuven have submitted a patent application (Sept. 15, 2017; EP 17191252.0.) on commercial use of the results.
Acknowledgments
We thank Teun Boekhout, Bart Theleen, and Raquel Quintilla Mateo (Westerdijk Fungal Biodiversity Institute, Utrecht) for help with initial characterization of the S. boulardii strains, Arne Claes for help with screening of the segregants, Lene Jespersen for her kind supply of S. boulardii strains, Gianni Liti for kind provision of the ARL and CIT strains, Renata Wicik for technical support, and Nico Vangoethem for help with preparation of the figures. This work was supported by an SBO grant (IWT 90043) from IWT-Flanders and the EC 7th Framework program (CORNUCOPIA project) to J.M.T. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions: J.M.T. and M.R.F.-M. conceived the project; B.O. and P.V. performed the experiments; B.O., P.V., M.R.F.-M, S.D.G., and J.M.T. analyzed the data; and B.O., M.R.F.-M, and J.M.T. wrote the manuscript.
Footnotes
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.243147.118.
-
Freely available online through the Genome Research Open Access option.
- Received August 17, 2018.
- Accepted June 20, 2019.
This article, published in Genome Research, is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.