Into the blue: Gene duplication and loss underlie color vision adaptations in a deep-sea chimaera, the elephant shark Callorhinchus milii
- Wayne L. Davies1,3,
- Livia S. Carvalho1,
- Boon-Hui Tay2,
- Sydney Brenner2,
- David M. Hunt1,4 and
- Byrappa Venkatesh2,4
- 1 UCL Institute of Ophthalmology, London EC1V 9EL, United Kingdom;
- 2 Institute of Molecular and Cell Biology, Agency for Science, Technology, and Research, Biopolis, Singapore 138673, Singapore
Abstract
The cartilaginous fishes reside at the base of the gnathostome lineage as the oldest extant group of jawed vertebrates. Recently, the genome of the elephant shark, Callorhinchus milii, a chimaerid holocephalan, has been sequenced and therefore becomes the first cartilaginous fish to be analyzed in this way. The chimaeras have been largely neglected and very little is known about the visual systems of these fishes. By searching the elephant shark genome, we have identified gene fragments encoding a rod visual pigment, Rh1, and three cone visual pigments, the middle wavelength-sensitive or Rh2 pigment, and two isoforms of the long wavelength-sensitive or LWS pigment, LWS1 and LWS2, but no evidence for the two short wavelength-sensitive cone classes, SWS1 and SWS2. Expression of these genes in the retina was confirmed by RT-PCR. Full-length coding sequences were used for in vitro expression and gave the following peak absorbances: Rh1 496 nm, Rh2 442 nm, LWS1 499 nm, and LWS2 548 nm. Unusually, therefore, for a deep-sea fish, the elephant shark possesses cone pigments and the potential for trichromacy. Compared with other vertebrates, the elephant shark Rh2 and LWS1 pigments are the shortest wavelength-shifted pigments of their respective classes known to date. The mechanisms for this are discussed and we provide experimental evidence that the elephant shark LWS1 pigment uses a novel tuning mechanism to achieve the short wavelength shift to 499 nm, which inactivates the chloride-binding site. Our findings have important implications for the present knowledge of color vision evolution in early vertebrates.
Early in vertebrate evolution, two major lineages emerged, the Agnatha or jawless vertebrates and the Gnathostomata or jawed vertebrates. The only extant members of the Agnatha are the hagfish and lampreys, whereas the Gnathostomata include all of the remaining extant vertebrate classes. The Chondrichthyes (cartilaginous fishes) reside at the base of the gnathostome lineage as the oldest extant jawed vertebrate group, where they share a common ancestor dating from at least 450 million years ago (Mya) (Sansom et al. 1996) with all other jawed vertebrates (i.e., bony vertebrates that include teleosts and tetrapods). Cartilaginous fishes are divided into two lineages, the holocephalans (chimaeras) and the elasmobranchs (sharks, rays, and skates), with the chimaeras diverging about 374 Mya (Cappetta et al. 1993) from the common chondrichthyan ancestor (Tudge 2000).
Daylight or photopic vision clearly arose in chordate evolution before the major split into jawless and jawed vertebrate lineages (Collin et al. 2003), but it remains uncertain whether jawless vertebrates possess dim light or scotopic vision (Pisani et al. 2006). This differential sensitivity to light is provided by the rod and cone photoreceptors within the classic duplex retina, with a single rod visual pigment responsible for scotopic vision and up to four different classes of cone pigments, each segregated into different clone classes, responsible for photopic and color vision. Cartilaginous fishes, like their teleost counterparts, occupy a wide range of biological niches. Traditionally, these fishes are thought to possess a poor visual system with eyes that are specialized for scotopic vision. These specializations include the presence of a tapetum at the rear of the eye for reflecting light back on to the photoreceptors, a high photoreceptor to ganglion cell summation ratio that increases sensitivity at the expense of acuity (Walls 1942), and in some species, an all-rod retina (Dowling and Ripps 1990; Ripps and Dowling 1990). However, duplex retinae with cone and rod photoreceptors have been identified in many elasmobranch species that include the catshark (Scyliorhinus spp.) (Neumayer 1897), the dogfish (Mustelus canis) (Schaper 1899), the lemon shark (Negaprion brevirostris) (Gruber et al. 1963; Cohen et al. 1990), the giant shovelnose ray (Rhinobatos typus) (Hart et al. 2004), the eastern shovelnose ray (Aptychotrema rostrata) (Hart et al. 2004), and the blue-spotted maskray (Dasyastis kuhlii) (Theiss et al. 2007).
Recently, it has been shown that a full complement of cone opsin genes is expressed in the retina of a jawless agnathan, the pouched southern-hemisphere lamprey Geotria australis (Collin et al. 2003), and that their expression is temporally regulated during the lamprey life cycle (Davies et al. 2007). The molecular basis for the visual pigments expressed in cartilaginous fishes remains, however, to be determined. Microspectrophotometry (MSP) has been used to characterize the spectral sensitivity of photoreceptors in a few species (Cohen et al. 1990; Hart et al. 2004; Theiss et al. 2007) and short, middle, and long wavelength-sensitive (LWS) cones have been identified. However, only the rod opsin sequence expressed in the retina of the little skate (Raja erinacea) (O'Brien et al. 1997), the dogfish (Galeus melastomus, accession no. Y17586), and the smaller-spotted catshark (Scyliorhinus canicula, accession no. Y17585) have been reported.
The genome of a chimaerid holocephalian, the elephant shark, Callorhinchus milii (also known as the ghost shark or elephant fish), has been sequenced and becomes, therefore, the first cartilaginous fish to be analyzed in this way (Venkatesh et al. 2007). As a group, the chimaeras have been largely neglected in favor of the more widespread and iconic sharks and batoids, and very little is known about the visual systems of these fishes. Unlike elasmobranchs that possess a large genome, the elephant shark has a compact genome that is better served for whole-genome sequencing (Venkatesh et al. 2005). Interestingly, comparisons between the genomes of the elephant shark and other species have identified many unexpected features, such as the conservation of ancient noncoding elements, a high degree of synteny, and the presence of specific ancient genes that have been lost in subsequent teleost and tetrapod radiations (Venkatesh et al. 2006, 2007).
Elephant sharks live on the continental shelves of southern Australia and New Zealand at depths between 200 and 500 m (Last and Stevens 1994). Adults mature at the age of 3–4 yr when they migrate into shallow bays and estuaries (6–30 m) during the warmer months to spawn. During this period, females lay two fertilized eggs per week on the sandy bottom. The eggs take 6–8 mo to hatch, and the juveniles then migrate to deeper waters. The elephant shark is therefore exposed to different light environments during its life cycle.
The aim of this study was to investigate the visual system of the elephant shark via a search of the elephant shark genome for visual pigment genes, to confirm the expression of each opsin in the retina, and to measure the spectral properties of each visual pigment so identified. In this way, the spectral sensitivity of the elephant shark visual system could be evaluated in relation to the ecology of its natural habitat.
Results
Scaffold sequences of the elephant shark visual pigment genes
Our search for opsin gene sequences in the elephant shark genome identified six scaffolds, of which two (accession nos. AAVX01025019.1 [2173 bp] and AAVX01065105.1 [3036 bp]) showed high identity to different coding regions of Rh1 (rod opsin class), two (AAVX01418754.1 [935 bp] and AAVX01124770.1 [1356 bp]) to different coding regions of Rh2 (green cone opsin class; MWS), and two (AAVX01110722.1 [1495 bp] and AAVX01158890.1 [1046 bp]) to coding regions of LWS (red cone opsin class; long wavelength sensitive, LWS). Oligonucleotides were designed that were specific to the exons on these scaffolds (Table 1) and the complete coding sequences of the genes were obtained by 5′- and/or 3′-RACE (rapid amplification of cDNA ends) using elephant shark eye/retina RACE-ready cDNA.
Oligonucleotide sequences used in 5′- and 3′-RACE and the generation of full-length recombinant C. milii opsin constructs for protein expression analysis
Elephant shark retinal rod opsin
Alignment of scaffolds AAVX01025019.1 and AAVX01065105.1 with known vertebrate Rh1 cDNA sequences showed high identity with exons 1 and 2 and 3–5, respectively, and inspection of the predicted exons demonstrated that all of the putative splice sites flanking these exons adhere to the GT-AG rule for exon–intron and intron–exon junctions (Mount 1982; see Table 2). These two scaffolds do not overlap, but a bacterial artificial chromosome (BAC) end sequence (BAC ID 068K20) (523 bp) was found that overlaps AAVX01025019.1 by 102 bp and AAVX01065105.1 by 296 bp. This enabled the full sequence of 3853 bp of the elephant shark Rh1 gene (from translation initiation to termination codon) to be determined. The gene consists of five exons (exons 1–5: 361, 169, 166, 240, and 129 bp) and four introns (intron 1 to intron 4: 900, 628, 630, and 630 bp). Using ESR1PEF and ESRIPER oligonucleotides (Table 1) and elephant shark retinal/eye cDNA, a full-length sequence of the Rh1 coding region of 1065 bp (accession no. EF565167) was PCR generated and confirmed by sequencing.
Exon–intron and intron–exon boundaries for Rh1 gene expressed in elephant shark retina
Cone opsins expressed in the elephant shark retina
Alignment of known vertebrate Rh2 cDNA sequences with scaffold sequences identified exon 2 (169 bp) in AAVX01418754.1 and exon 3 (166 bp) in AAVX01124770.1 of the elephant shark Rh2 gene. Sense (ESR2F1 and ESR2F2) and antisense (ESR2R1 and ESR2R2) oligonucleotides (Table 1) were used in 5′- and 3′-RACE amplifications with eye/retinal cDNA; sequencing of the PCR products confirmed that these two Rh2 exons are part of a single cDNA encoding an Rh2 opsin (1068 bp; accession no. EF565168).
A similar screening of the elephant shark genome with LWS opsin sequences revealed two positive scaffolds, AAVX01158890.1 containing exon 3 (169 bp), and AAVX01110722.1 containing a partial sequence for exon 5 (202 bp of a predicted 240 bp) and complete sequences for intron 5 (257 bp) and exon 6 (117 bp). Initial PCRs using either gDNA or eye/retinal cDNA as template and oligonucleotides designed specifically to each predicted LWS exon failed to amplify a sequence that linked exon 3 in AAVX01158890.1 to exons 5 and 6 in AAVX01110722.1 (data not shown). Various scaffold-specific sense and antisense RACE oligonucleotides (ESL1F1, ESL1F2, ESL2F1, ESL2F2, and ESL1R1, ESL1R2, ESL2R1, and ESL2R2 in Table 1) were therefore used to generate the 5′- and 3′-ends extending from each predicted LWS exon. Finally, PCR amplification with eye/retinal cDNA and sequencing confirmed that the elephant shark genome possesses two distinct LWS genes, with exon 3 in AAVX01158890.1 belonging to LWS2 (coding region of 1095 bp, accession no. EF565166) and exons 5 and 6 in AAVX01110722.1 to LWS1 (coding region of 1098 bp, accession no. EF565165). 5′- and 3′-RACE amplifications with eye/retinal cDNA confirmed that both genes are expressed in the retina. Primers specific for each LWS isoform (Table 1) were then used to amplify full-length coding sequences; the sequences of these amplicons confirmed the presence of two distinct LWS sequences that share a percentage identity of 77% and 79% at the nucleotide and amino acid level, respectively.
Finally, screening with the two remaining vertebrate opsin genes, short wavelength-sensitive type 1 (SWS1) and short wavelength-sensitive type 2 (SWS2), failed to identify any positive scaffolds. Three degenerate oligonucleotides (SWSF, SWS1R, and SWS2R in Table 1), designed to conserved regions of the SWS1 and SWS2 genes, were used to probe for expression of either gene in the elephant shark retina. All oligonucleotide pairs amplified the fragments of the expected sizes from Fugu eye cDNA (positive control), but not from the elephant shark retinal cDNA. In the human genome, the SWS1 gene, currently known as OPN1SW, is only 3.3 kb from the calumenin precursor (CALU) gene on the upstream side, with NAG6 immediately downstream. The zebrafish orthologs of CALU and SWS1 (opn1sw1) are similarly linked with a 3.1-kb intergenic region, but the gene encoding Transportin-3 (Tnpo3) is located immediately downstream of zebrafish Sws1 gene. The 1.4× coverage assembly of the elephant shark, which represents about 75% of the genome, did not contain any sequences for SWS1. By applying a TBLASTN search, however, we identified a scaffold that contained an elephant shark ortholog of the CALU gene. PCR oligonucleotides specific for the elephant shark Calu gene were designed and used to screen an elephant shark BAC library. A single BAC (#51A04) containing the Calu sequence was identified and DNA from this BAC was sequenced, identifying 92 contigs, which, when assembled, gave a combined length of 228 kb. A BLASTX search of these contigs showed that the assembly does not contain sequences for SWS1 but contained orthologous sequences for elephant shark CALU and TNPO3 genes. One contig of about 7.8 kb contains the last exon for CALU with 4.6-kb downstream sequence. Synteny analysis showed, therefore, that the SWS1 gene has been lost from the elephant shark genome. A similar “gene-mining” approach was used to search for an SWS2 ortholog in the elephant shark genome. In vertebrates, such as the zebrafish, the genes for host cell factor C1 (Hcfc1) and LWS opsin flank, respectively, the upstream and downstream sides of the SWS2 gene. While a BLASTX search of the elephant shark genome failed to identify an ortholog of SWS2, contigs with identity to both HCFC1 and LWS genes were detected, suggesting the absence of SWS2 from the elephant shark genome. Thus, both PCR methods and bioinformatics consistently show that the genome of the elephant shark lacks an ortholog of the SWS2 gene.
Phylogeny of elephant shark opsin genes
Elephant shark Rh1, Rh2, LWS1, and LWS2 nucleotide sequences were subjected to phylogenetic analysis. Nucleotide sequences were preferred to amino acid sequences, as phylogenetic information can be lost with the latter. In order to obtain a consensus for the evolutionary relationships over the long time period involved, three mathematically distinct methods of phylogenetic analysis (neighbor-joining [NJ], maximum likelihood [ML], and Bayesian probability [BP]) were used. All three methods placed each elephant shark opsin into the expected opsin clade with high-probability scores, thereby providing confirmation that these sequences are orthologs of the pigment genes of other vertebrates (Fig. 1). In particular, LWS1 and LWS2 clade together with a probability score of >94, verifying that these two sequences are very closely related and likely to be the result of a gene duplication. All four elephant shark opsins were positioned ancestrally to their teleost counterparts within each clade by all three phylogenetic methods. Also as expected, elephant shark Rh1 and Rh2 sequences were positioned more recently than lamprey RhA (sea, arctic, and pouched lampreys) and RhB opsins (pouched lamprey only). In contrast however, all three methods placed the elephant shark LWS1 and LWS2 genes ancestral to the LWS opsin of the pouched lamprey, suggesting that lamprey and elephant shark LWS opsins differ in their relative rates of evolution. This was confirmed by the application of Tajima's relative rate test (Tajima 1993; Table 3). When zebrafish and goldfish LWS (opn1lw1) sequences were each compared separately with the LWS1 and LWS2 sequences of the elephant shark and with each other, the relative rates of nucleotide substitution appear to be very similar, indicating that each gene is under the same evolutionary pressure, whereas comparisons of the pouched lamprey LWS sequence with those of the zebrafish, goldfish, and elephant shark showed that the relative rate of nucleotide substitution in the lamprey sequence is significantly different from the other four species, thereby accounting for its “misplaced” phylogenetic position.
Phylogenetic analysis of LWS1, LWS2, Rh2, and Rh1 elephant shark vertebrate retinal opsin sequences and coding sequences of other related vertebrate species. (A) A neighbor-joining tree was constructed (Saitou and Nei 1987) by applying a Kimura 2-parameter substitution matrix (Kimura 1980) with 1000 bootstrapping replications (degree of support for internal branching is shown as a percentage at the base of each node). (B) A maxiumum likelihood tree was generated with a Kimura 2-parameter substitution matrix (Kimura 1980), a gamma distribution parameter of 1 using PHYML (Guindon et al. 2005) and bootstrapping with 500 replicates (degree of support for internal branching is shown as a percentage at the base of each node). (C) A Bayesian probabilistic inference method was performed with a Metropolis Markov chain Monte Carlo algorithm (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). A general time-reversal model (Lanave et al. 1984) was used with posterior probability values (represented as a percentage) indicated at the base of each node. The scale bar indicates the number of nucleotide substitutions per site. The Drosophila melanogaster (fruit fly) Rh4 (accession no. NM057353) was used as an outgroup. The sequences used for generating the tree are as follows: (a) RhA/Rh1 opsin class: zebrafish (Danio rerio), NM131084; goldfish (Carassius auratus), L11863; guppy (Poecilia reticulata), Y11147; John Dory (Zeus faber), Y14484; smaller spotted catshark (Scyliorhinus canicula), Y17585; dogfish (Galeus melastomus), Y17586; little skate (Raja erinacea), U81514; elephant shark (Callorhinchus milii), EF565167; sea lamprey (Petromyzon marinus) (RhA), AH005459; Arctic lamprey (Lethenteron japonica) (RhA), M63632; pouched lamprey (Geotria australis) (RhA), AY366493; (b) RhB/Rh2 opsin class: zebrafish (Danio rerio), AB087805 (Rh2.1), AB087806 (Rh2.2), AB087807 (Rh2.3), AB087808 (Rh2.4); goldfish (Carassius auratus), L11865 (Rh2.1), L11866 (Rh2.2); bluefin killifish (Lucania goodei), AY296739; rainbow trout (Oncorhynchus mykiss), AF425072; elephant shark (Callorhinchus milii), EF565168; pouched lamprey (Geotria australis) (RhB), AY366494; (c) SWS2 opsin class: zebrafish (Danio rerio), NM131192; goldfish (Carassius auratus), L11864; bluefin killifish (Lucania goodei), AY296737; rainbow trout (Oncorhynchus mykiss), AF425075; pouched lamprey (Geotria australis), AY366492; (d) SWS1 opsin class: zebrafish (Danio rerio), NM131319; goldfish (Carassius auratus), D85863; pouched lamprey (Geotria australis), AY366495; (e) LWS opsin class: zebrafish (Danio rerio), NM131175; goldfish (Carassius auratus), L11867; pouched lamprey (Geotria australis), AY366491; elephant shark (Callorhinchus milii), EF565165 (LWS1), EF565166 (LWS2).
Tajima's relative rate tests on paired LWS sequences using a χ2 distribution and one degree of freedom, with the fruit fly Rh4 sequence as an outgroup
Evolutionary analysis of the Rh1 gene family, using NJ, ML, and BP methods, places the elephant shark sequence basal to the elasmobranchs, with the little skate (R. erinacea) more ancestral than the smaller-spotted catshark (Scyliorhinus canicula) and the dogfish (Galeus melastomus) (Fig. 1). This is consistent with other molecular phylogenetic analyses of chondrichthyan fishes (Douady et al. 2003; Mallatt and Winchell 2007).
Except for the positioning of the RhA and RhB genes of the pouched lamprey, the branching patterns of the phylogenetic trees generated by NJ, ML, and BP methods are essentially identical (Fig. 1). In the NJ-tree, the lamprey RhA and RhB genes clade together as a sister group to the Rh1 opsin genes of the chondrichthyes and teleosts (Fig. 1A). This suggests that the Rh1 and Rh2 gene duplication occurred in the gnathostome lineage only with an independent duplication of an ancestral Rh opsin gene to give RhA and RhB in the Agnathans (Collin et al. 2003). Alternatively, the Rh1/Rh2 duplication may have occurred prior to the divergence of separate jawless and jawed vertebrate lineages, with a loss of the lamprey Rh2 gene and an independent gene duplication taking place in the lamprey lineage to give the RhA and RhB gene variants. Conversely, both the ML (Fig. 1B) and BP (Fig. 1C) methods place the lamprey RhA genes with the gnathostome Rh1 group and the pouched lamprey RhB gene with the Rh2 opsin class, with both the lamprey Rh genes ancestral to the Rh1 and Rh2 genes of the elephant shark. The positioning of the lamprey RhA and RhB genes with the gnathostome Rh1 and Rh2 genes, respectively, is consistent with the analysis of Pisani et al. (2006).
Spectral sensitivity of elephant shark visual pigments
Full-length coding sequences for elephant shark Rh1, Rh2, LWS1, and LWS2 opsins were generated by PCR as single amplicons using protein expression sense (ESR1PEF, ESR2PEF, ESL1PEF, and ESL2PEF) and antisense (ESR1PER, ESR2PER, ESL1PER, and ESL2PER) oligonucleotides (Table 1) and expressed in vitro. Each recombinant pigment protein was harvested, reconstituted with 11-cis-retinal, and subjected to spectrophotometric analysis. Dark spectra for each visual pigment were determined across a wide range of wavelengths (250–800 nm), followed by light bleaching under a fluorescent lamp for 1 h. Peak spectral sensitivity values (λmax) were obtained by fitting a Govardovskii template (Govardovskii et al. 2000) to the difference spectra (dark minus bleach). As shown in Figure 2, all four elephant shark pigments were successfully regenerated. The Rh1 pigment gave a λmax value of 496 ± 0.1 nm (Fig. 2A), which is similar to the peak spectral sensitivity of around 500 nm found for many other vertebrate rod photoreceptors (Yokoyama 2000), whereas the Rh2 pigment gave a λmax value of 441 ± 1.0 nm (Fig. 2B), which is substantially more short wavelength shifted than any other Rh2 pigment reported so far. Regeneration of LWS1 and LWS2 opsins with 11-cis-retinal produced pigments with λmax values of 499 ± 0.3 nm and 548 ± 2.2 nm, respectively (Fig. 2B). The elephant shark LWS2 visual pigment peaks in the yellow to red range of the visible spectrum, which is similar to LWS pigments in other species (Yokoyama 2000). The peak spectral sensitivity of the LWS1 pigment, however, is short wavelength shifted into the blue–green or middle-wavelength-sensitive (MWS) range of the visible spectrum. With a λmax value 9 nm, more short wavelength shifted than the mouse green-sensitive LWS pigment (λmax = 508 nm) (Sun et al. 1997), the elephant shark LWS1 visual pigment is the most short wavelength shifted pigment known for the LWS class.
Absorption spectra of regenerated C. milii (A) rod (Rh1) and (B) cone (Rh2, LWS1, and LWS2) visual pigments. For Rh1 pigments, representative dark (●) and bleached spectra (○) are shown, with difference spectra that have been fitted with a Govardovskii et al. (2000) template (line) in the inset to determine the λmax value. For Rh2, LWS1, and LWS2 pigments, representative difference spectra are shown that were fitted to Govardovskii et al. (2000) templates to determine the λmax values. For LWS1 pigments, the spectral peak of absorbance was determined in the presence (continuous line) and absence of chloride ions (dotted line).
Putative amino acids involved in the spectral tuning of elephant shark visual pigments
Amino acid alignments of elephant shark Rh1, Rh2, LWS1, and LWS2 opsin sequences together with the bovine Rh1 rod opsin sequence (accession no. NP001014890) are shown in Figure 3. Note that for all of the elephant shark opsins, the critical residues involved in maintaining the function and structural integrity of the pigment are conserved.
Alignment of the amino acid sequences of visual pigments expressed in the elephant shark (Callorhinchus milii; CM) and the common cow (Bos taurus, BT). (Rh1) Rod opsin; (LWS1, LWS2) long-wavelength-sensitive 1 and 2; (Rh2) cone rhodopsin 2 (middle-wavelength-sensitive). (*) An identical consensus residue between all four elephant shark opsin sequences and bovine Rh1. (: and .) A conserved or semiconservative amino substitution, respectively, with the codon-matched protein alignment. Gaps inserted to maintain a high degree of identity present between the opsin sequences derived from C. milii retina and bovine Rh1 are indicated by dashes (–). Seven putative transmembrane domains (TMDs) are indicated by gray shading. The TMDs shown for bovine rhodopsin were determined by crystallography (Palczewski et al. 2000). The putative positions of the TMDs for each elephant shark visual pigment were predicted online using TMHMM Server Version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Residues identified as being critical for correct opsin protein conformation (boxed). Comparison of the opsin amino acid sequences of C. milii and bovine rod opsin demonstrated that the critical residues involved in the maintenance of the tertiary structure of the opsin molecule are present. With the use of the conventional numbering system of the bovine rod opsin polypetide sequence, these key sites include (1) three conserved cysteine (C) residues at positions 110 (TMD3), 185 (ECD2), and 187 (ECD2) that are involved in disulphide bond formation (Karnik and Khorana 1990), except for a Thr (T) residue at position 185 in the elephant shark LWS1 and LWS2 opsins, which are also conserved throughout the rest of the vertebrate LWS opsin class; (2) a conserved glutamate (E) at position 113 (TMD3) that provides the negative counterion to the proton of the Schiff base (Sakmar et al. 1989); (3) a conserved glutamate (E) at position 134 (TM3) that provides a negative charge to stabilize the inactive opsin molecule (Cohen et al. 1993); (4) a conserved lysine (K) at position 296 (TM7) that is covalently linked to the chromophore via a Schiff base (Dratz and Hargrave 1983); (5) conservation of two cysteine (C) residues at putative palmitoylation positions 322 and 323 (Ovchinnikov et al. 1988) in both elephant shark Rh1 and Rh2 opsins, but not LWS1 and LWS2 opsins; (6) the presence of a number of Ser (S) and Thr (T) residues in the carboxyl terminus, which are potential targets for phosphorylation by rhodopsin kinases in the deactivation of metarhodopsin II (Palczewski et al. 1993; Sakmar and Fahmy 1996; Zhao et al. 1997); and (7) the conserved glycosylation sites at positions 2 and 15 (Sakmar and Fahmy 1996) in the MWS opsins (Rh1 and Rh2) identified in the retina of C. milii.
To investigate the mechanisms that underlie the spectral tuning of each visual pigment expressed in the elephant shark retina, the residues at key sites known to be critical for determining the λmax value of the pigment class were identified (Fig. 3; Table 4). The residues at seven sites, 83, 122, 207, 211, 265, 292, and 295, are thought to modulate the spectral sensitivities of Rh1 and Rh2 visual pigments (Yokoyama 2000). These tuning sites in ancestral Rh1 and Rh2 pigments (Davies et al. 2007) differ only at position 295 with Ala in Rh1 and Ser in Rh2 (Table 4, top). The tuning sites of the elephant shark Rh1 sequence are identical to the ancestral sequence, and the λmax of the elephant shark Rh1 pigment at 496 nm is similar to that of many other vertebrate species (Yokoyama 2000).
Spectral tuning of elephant shark visual pigments
The Rh2 pigment of the elephant shark possesses Ser295, which would be expected from site-directed mutagenesis experiments with bovine Rh1 (Lin et al. 1998) to generate a 5-nm short wavelength shift in λmax. A further four substitutions (Asn83/Gln122/Leu207/Ser292) compared with the ancestral Rh2 opsin sequence are present at tuning sites (Table 4, top), and site-directed mutagenesis at these sites is again known to cause changes in λmax. For example, in the Rh1 pigments of bovine and some deep-sea teleosts, an Asp83Asn substitution results in a short wavelength shift of 6 nm (Yokoyama et al. 1999; Hunt et al. 2001), a Glu122Gln substitution causes a large short wavelength shift of 19 nm in bovine Rh1 (Nathans 1990), and Leu207Met and Ser292Ala changes in the Rh2 pigment of the coelacanth cause a 6 and 8-nm long wavelength shift, respectively (Yokoyama et al. 1999). If additive, these four amino acid substitutions would generate a net short wavelength shift of 39 nm, which would generate most of the shift from 495 nm in the ancestral Rh2 pigment (Table 4, top). The additional shift of 14 to 445 nm must be due to other novel tuning sites.
For the LWS pigments, residues at five sites (Ser164, His181, Tyr261, Thr269, and Ala292) have been previously shown to locate the peak spectral sensitivity of the putative ancestral LWS pigment at 560 nm (Yokoyama and Radlwimmer 1999). The elephant shark LWS2 pigment differs at only one position with Phe261 rather than Tyr261. This substitution is known to short wavelength shift the λmax by 7 nm (Yokoyama and Radlwimmer 1999), which would give a predicted λmax value of around 553 nm for the elephant shark LWS2 pigment (Table 4, bottom); the value of 548 nm obtained for the in vitro-expressed pigment is within a few nanometers of this predicted value. The LWS1 pigment differs from the ancestral complement of tuning residues at three positions (Phe261, Ala269, and Ser292) (Table 4, bottom). Substitution at each of these sites is known to short wavelength shift the λmax value of LWS pigments by 7, 15, and 16 nm, respectively (Yokoyama and Radlwimmer 1999), and, if additive, would give a combined shift of 38 nm to a predicted value of 522 nm (Table 4, bottom). The λmax value for the in vitro-expressed pigment was, however, at 499 nm, which is 23 nm more short wavelength shifted than predicted.
Anion sensitivity of LWS pigments
Many LWS pigments are anion sensitive (Kleinschmidt and Harosi, 1992), with His181 and Lys184 forming a chloride-binding site (Wang et al. 1993). Replacement of His181, as has occurred naturally in the mouse LWS pigment, abolishes chloride binding and generates a 28-nm short wavelength shift in λmax (Sun et al. 1997). The elephant shark LWS1 pigment still possesses His181 and therefore would be expected to bind chloride ions, so regeneration of the pigment in vitro in the absence of chloride ions would be expected to give a further short wavelength shift in λmax. Regeneration in the absence of chloride ions (Fig. 2B) showed, however, that the λmax of the pigment is almost identical at 494 ± 2.6 nm, indicating that despite the presence of His181, the pigment is not responsive to chloride ion binding.
Residue differences between the LWS1 and LWS2 pigments at spectral tuning sites are restricted to changes at positions 269 and 292. Previous studies of the former site using site-directed mutagenesis to substitute residues gave spectral shifts of only 15 nm (Yokoyama and Radlwimmer 1999), and thus cannot fully account for the peak absorbance differences between LWS1 and LWS2 pigments of the elephant shark. In contrast, amino acid differences at position 292 have been shown to result in spectral shifts that are greater than the expected values (Fasick et al. 1998), so it is possible that the residue (Ala or Ser) present at site 292 affects chloride binding at His181. To investigate this, a series of single and double mutants was generated and their spectral sensitivities determined (Fig. 4). A His181Tyr mutation in the LWS2 pigment gave a 28-nm short wavelength shift to a λmax of 521 ± 2.0 nm, which is identical to the shift observed in other studies. This clearly shows that the chloride-binding site is functional in the LWS2 pigment. Conversely, the same mutation in the LWS1 pigment (with a λmax of 498 ± 0.3 nm) failed to short wavelength shift the λmax, demonstrating that the loss of a functional chloride-binding site does not affect spectral tuning. This implies that the chloride-binding site is already inactive in this pigment, despite the presence of His181. Introduction of an Ala292Ser mutation into the LWS2 pigment gave a λmax value of 504 ± 1.0 nm, which is short wavelength shifted by 44 nm compared with the wild-type pigment and significantly greater than the predicted shift of 16 nm for a mutation of this kind (Yokoyama and Radlwimmer 1999). If additive, both His181Tyr and Ala292Ser mutations in the LWS2 pigment would be expected to short wavelength shift the λmax to 473 nm, whereas the observed peak for the double mutant was at 510 ± 0.2 nm, very similar to the single Ala292Ser mutant (with a λmax of 504 ± 1.0 nm). This demonstrates that the presence of Ser292 is sufficient to render the chloride-binding site inactive in the elephant shark LWS2 pigment.
Absorption spectra of regenerated C. milii LWS1 (left) and LWS2 (right) mutants. (Top) His181Tyr single substitutions in both LWS1 and LWS2 pigments. (Middle) Ser292Ala (LWS1) or Ala292Ser (LWS2) single substitutions. (Bottom) His181Tyr and Ser292Ala (LWS1) or His181Tyr and Ala292Ser (LWS2) double substitutions. For all pigments, representative difference spectra are shown that were fitted to Govardovskii et al. (2000) templates to determine the λmax values. The vertical dotted line shows the relative position of the spectral peak values for both wild-type LWS1 (499 nm) and LWS2 (548 nm) regenerated pigments.
Since the His181Tyr mutation has already established that an inactive chloride-binding site is present in the LWS1 pigment, a His181Tyr and Ser292Ala double mutant would be expected to result in a spectral shift that was solely dependent on the Ser292Ala change. When these mutations were simultaneously introduced into the LWS1 pigment, an 11-nm long wavelength shift was obtained to a λmax value of 510 ± 0.5 nm, similar, therefore, to the predicted 16-nm long wavelength shift for Ser292Ala mutations (Yokoyama and Radlwimmer 1999). When only the single Ser292Ala mutation was introduced, thereby leaving the chloride-binding site intact with His181, the λmax was long wavelength shifted by 35 to 534 ± 1.0 nm. This corresponds to the expected shift for chloride binding; it would appear, therefore, that a Ser292Ala substitution enables chloride binding to occur. Collectively, these data strongly support our hypothesis that a Ser residue at site 292 is responsible for inactivating the chloride binding site.
Discussion
By combining data mining of the elephant shark genome with gene amplification and sequencing, we have been able to identify three cone visual pigment genes, one belonging to the Rh2 opsin class and two belonging to the LWS opsin class, as well as an Rh1 rod opsin gene. We have obtained full-length sequences for all four genes and demonstrated by RT–PCR that all four are expressed in the retina of the elephant shark. In contrast, no evidence for the presence of either an SWS1 or SWS2 gene was obtained. This is the first study to identify cone opsin genes in any cartilaginous fish.
The two LWS genes, LWS1 and LWS2, have presumably arisen from a gene duplication. LWS gene duplications have also been reported in a number of species of teleost fish (Yokoyama and Yokoyama 1990; Register et al. 1994; Chinen et al. 2003; Fuller et al. 2004; Kasai and Oshima 2006; Matsumoto et al. 2006; Weadick and Chang 2007), in Old World primates (Nathans et al. 1986; Ibbotson et al. 1992), and in a single species of New World primate, the howler monkey (Dulai et al. 1999). In primates, the duplication is directly linked to the attainment of trichromacy, and the same may be the case for the elephant shark. In the elephant shark, however, the third pigment is encoded by an Rh2 gene, not an SWS1 gene as in primates. Whether the duplication of the LWS gene in the elephant shark is unique to the chimaerid lineage or occurred in the chondrichthyan ancestor remains to be determined. Furthermore, gene duplications such as the two LWS genes in primates (Nathans et al. 1986; Ibbotson et al. 1992; Dulai et al. 1999) and the four Rh2 genes in zebrafish (Tsujimura et al. 2007) lie in an array under the control of a local control region (LCR) that results in exclusive expression of a single opsin in a particular photoreceptor. Whether the LWS1 and LWS2 genes of the elephant shark also lie in an array and are under the control of an LCR cannot be ascertained from the current build of the elephant shark genome.
The elephant shark Rh1 gene is ∼3.9 kb in length and comprises five exons and four intervening introns. The average length of the introns is short at ∼700 bp, consistent, therefore, with the overall compact size of the elephant shark genome (Venkatesh et al. 2005). In contrast, much larger introns have been identified in the partial sequence of the Rh1 gene of the little skate (O'Brien et al. 1997), whereas in all teleosts studied so far, the Rh1 gene totally lacks introns. Since intron loss is a very rare event, it most probably arose as a single retrotransposition event of a processed mRNA (Fitzgibbon et al. 1995; Bellingham et al. 2003). The five exon/four intron structure of the Rh1 gene in the elephant shark and in other nonteleost vertebrates demonstrates that this intron loss is restricted to teleosts.
Phylogenetically, each elephant shark opsin is orthologous to a class of opsin previously identified in other vertebrates. Close inspection of Rh1 and Rh2 gene families demonstrates that the elephant shark Rh1 and Rh2 genes are consistently located ancestral to their teleost counterparts, irrespective of the phylogenetic method used. “True” members of the vertebrate Rh1 and Rh2 opsin classes were present, therefore, in the holocephalans (Douady et al. 2003; Mallatt and Winchell 2007). Holocephalans shared a common ancestor with the elasmobranchs around 370 Mya; Rh1 and Rh2 opsins were already present, therefore, in the common ancestor of jawed vertebrates.
When expressed in vitro and regenerated with 11-cis-retinal, λmax values of 442, 499, and 548 nm were obtained for the elephant shark Rh2, LWS1, and LWS2 cone visual pigments, respectively, and a λmax of 496 nm for the rod (Rh1) pigment. The λmax of the Rh1 pigment is similar to that found for the rod pigments of many other vertebrate species. Moreover, the residues present at positions 83, 122, 207, 211, 265, 292, and 295, the key sites for spectral tuning, are identical to the deduced sequence of the ancestral Rh1 opsin (Davies et al. 2007) and therefore account for the spectral location of the λmax. In contrast, all three cone pigments are short wavelength shifted with respect to the λmax values predicted from residues present at key tuning sites (Yokoyama 2000), indicating that other amino acid substitutions must effect tuning. The Rh2 pigment is 10 nm more short wavelength shifted than the Rh2-A pigment of Orysias latipes (medaka) at 452 nm (Matsumoto et al. 2006) and 25 nm more short wavelength shifted than the Rh2 pigment of Gekko gekko (Tokay gecko) at 467 nm (Takenaka and Yokoyama 2007). The elephant shark Rh2 visual pigment is therefore the most short wavelength shifted pigment of this class thus far reported.
The λmax of the elephant shark LWS2 pigment at 548 nm is consistent with the amino acid residues present at the major tuning sites for LWS pigments (Yokoyama and Radlwimmer 1999), whereas the λmax of the LWS1 pigment at 499 nm is substantially more short wavelength shifted than can be explained by substitution at these sites. Unique to LWS pigments, the spectral peak is long wavelength shifted by the binding of chloride ions to the opsin protein via a chloride-binding site formed by His181 and Lys184 (Wang et al. 1993). Loss of this site by replacement of His181 with Tyr, as seen in the native mouse LWS pigment, generates a 28-nm short wavelength shift to a λmax of 508 nm (Sun et al. 1997). The elephant shark LWS1 pigment, although retaining His181, is further shifted to 499 nm. Nevertheless, our site-directed mutagenesis and in vitro pigment regeneration experiments indicate that this short wavelength shift is also achieved by the inactivation of the chloride-binding site, not by mutation of the site itself, but by interaction with the Ser292. The predicted positions of the known tuning sites for LWS pigments were obtained from the three-dimensional crystal structure of bovine rhodopsin (Palczewski et al. 2000). This places residues 181 and 292 in close proximity to each other and adjacent to the Schiff base that links the retinylidene chromophore to the opsin protein via a covalent bond at Lys296 (Fig. 5). In contrast, the other major LWS tuning sites at positions 164, 261, and 269 are located distally toward the β-ionone ring of the chromophore. The residues that cause the largest shifts in the spectral peak (i.e., 181 and 292) are therefore located near the Schiff base, with those that result in smaller changes in λmax located closer to the β-ionone ring. Whether the inactivation of the chloride-binding site by Ser292 is caused directly by steric inhibition by the side chain of the residue present at 292 or indirectly by changing the overall conformation of the visual pigment, and thus access of chloride ions to the retinal binding pocket, remains to be determined. The model that we propose (Fig. 5) is based on the intrinsic charge properties of the amino acid side chains and involves a negatively charged chloride ion acting as a counterion to the positive charge of the His181, but only when Ala is present at site 292. When His181 is replaced with Tyr181, the positive charge is lost and chloride binding is abolished irrespective of the residue present at site 292. However, when both His181 and Ser292 are present, as in the elephant shark LWS1 pigment, we propose that the negative polarity of the hydroxyl group of Ser292 acts either to repulse the negatively charged chloride ion or itself provides a “counterion” to the positively charged His181. The only other LWS pigments so far reported that pair His181 with Ser292 are found in three deeper-dwelling cetaceans, the bottlenose dolphin (Tursiops truncates), the pilot whale (Globicephala melas), and the harbor porpoise (Phocoena phocoena) (Newman and Robinson 2005). In the bottlenose dolphin pigment, a Ser292Ala substitution causes a long wavelength shift from wild-type λmax of 524 to 552 nm, a change of 28 nm (Fasick et al. 1998), which is almost identical to the 33-nm long wavelength shift observed when a Ser292Ala substitution in introduced into the LWS1 visual pigment of the elephant shark. Thus, the presence of Ser292 in the LWS pigments of all of the deeper-dwelling cetaceans identified so far may act to inactivate the chloride-binding site.
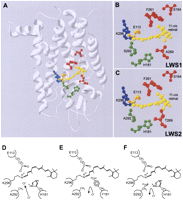
(A) Structural model of elephant shark LWS visual pigments showing the relative position of the five key LWS tuning sites (164, 181, 261, 269, and 292) within the retinal-binding pocket containing the retinylidene chromophore (yellow), the Lys296 chromophore attachment site via a Schiff base (blue), and the Glu113 counterion (orange). LWS tuning sites are color coded whether they are situated proximally to (green) or distally (red) from the Schiff base. Enlarged view showing the key residues of (B) LWS1 and (C) LWS2 pigments surrounding the retinylidene chromophore. The model was created using Swiss Model (Guex and Peitsch 1997) and is based on the crystal structure of bovine rhodopsin (Palczewski et al. 2000). A schematic representation of the proposed model for the interaction between residues 181 and 292 in close proximity to the Schiff base of the chromophore, showing (D) a chloride ion acting as a counterion to the positive charge present on His181, (E) the loss of binding and repulsion of a chloride ion due to the net negative polar charge of the Tyr residue at position 181, and (F) the net negative polar charge of the hydroxyl side chain of Ser292 repelling the negatively charged chloride ion and acting as a counterion to the positive charge present on His181. In all cases, the Glu113 counterion and Schiff base linking Lys296 and the retinylidene chromophore are shown. Putative electrostatic attractions between amino acid side chains and ions are highlighted by a dotted ellipse.
Short wavelength shifts in the peak absorbances of visual pigments are a relatively common adaptation of the visual system of deep water fish (Partridge et al. 1989; Bowmaker et al. 1994; Hunt et al. 2001; Sugawara et al. 2005). Down-welling sunlight penetrates the clearest oceanic water to a maximum of 1000 m (Jerlov 1976) and shows increasing attenuation at shorter and longer wavelengths with increasing depth. Pigments are therefore tuned to match the peak transmission around 480 nm. In most such cases, such adaptations are restricted to rod pigments, as most deep-water species have dispensed with cones. However, members of the species flock of cottoid fish in Lake Baikal that are adapted to increasing depths have not only short wavelength shifted their rod pigments, but have retained cones with Rh2 (Bowmaker et al. 1994; Hunt et al. 1996) and SWS2 pigments (Cowing et al. 2002) that are also short wavelength shifted. The short wavelength shifts seen in the cone pigments of the elephant shark may also be an adaptation, therefore, to light quality at depth in the ocean as it occupies a depth range of 200–500 m, although this is not seen for the rod pigment that retains a peak sensitivity at 496 nm as found in many shallow water fish and terrestrial vertebrates.
The coelacanth that lives at depths of 200 m possesses only Rh1 and Rh2 genes, with both pigments short wavelength shifted (Yokoyama et al. 1999). It has therefore lost a cone-based color vision system, a function that has been retained in the elephant shark that resides at comparable depths. Unlike the coelacanth, however, the adult elephant shark migrates into shallower estuarine waters to spawn. The novel form of trichromacy that the retention and duplication of the LWS gene is likely to provide may therefore be an adaptation to the varying light environment of the deep and shallow waters encountered by the elephant shark.
Methods
Tissues and RNA
The elephant shark tissues were collected at Queenscliff, Victoria, Australia from fresh shallow-caught specimens using recreational fishing equipment, and frozen immediately in liquid nitrogen. The frozen tissues were stored at −80°C. Total RNA was extracted from eye/retinal tissue using the TRIzol Reagent from Invitrogen.
Rapid amplification of cDNA ends (RACE)
Single-strand 5′- and 3′-RACE-ready cDNA was prepared from 1 μg of total RNA using the SMART RACE cDNA Amplification kit. Then, 5′- and 3′-ends of cDNA were amplified in a nested PCR reaction according to the manufacturer's instructions. The first-round PCR was carried out with a universal oligonucleotide (supplied in the SMART RACE cDNA Amplification kit) and a sense (3′-RACE, F1) or antisense (5′-RACE, R1) gene-specific oligonucleotide (Table 1), using single-strand cDNA as a template. A nested universal oligonucleotide (supplied in the SMART RACE cDNA Amplification kit) and a sense (3′-RACE, F2) or antisense (5′-RACE, R2) gene-specific oligonucleotide (Table 1) were used in a second-round PCR with 1 μL of 1:20 dilution of the first-round PCR reaction mixture as a template. The product of the nested PCR was run on an agarose gel, excised from the gel, purified, and sequenced either directly or after cloning into a pGEM-T vector (Promega). Sequencing was done using the dye-terminator technology on an ABI 3730xl DNA analyzer.
Elephant shark scaffolds for visual pigment genes
The 1.4× coverage elephant shark sequences (http://esharkgenome.imcb.a-star.edu.sg/), comprising 640,131 scaffolds, were searched for visual pigment genes using the TBLASTN algorithm, and human and zebrafish visual pigment gene protein sequences as bait. The scaffold sequences that showed a good homology (e-value cutoff at 1 × 10−7) to the query sequences were searched against the nonredundant protein database at NCBI (http://www.ncbi.nlm.nih.gov/) using BLASTX algorithm, and those that showed homology with a visual pigment gene were retained. All of the identified scaffolds contained only fragments of visual pigment genes. Full-length coding sequences of the genes were obtained by 5′- and 3′-RACE using oligonucleotides specific to the exonic sequences. Where required, similar genome searches were used to detect genes syntenic with those encoding visual pigments.
PCR for SWS genes in elephant shark
Sequences for SWS1 and SWS2 proteins from various vertebrates including lamprey were aligned using ClustalW to identify highly conserved regions. The following degenerate oligonucleotides corresponding to the highly conserved sequences were designed: SWSF: 5′-TGYGGNCCNGAYTGGTAYAC-3′ (corresponding to the amino acid stretch CGPDWYT conserved in both SWS1s and SWS2s); SWS1R: 5′-GTNGCNGAYTCYTGYTGYTG-3′ (complementary to amino acid sequence QQQESAT conserved in SWS1s); and SWS2R: 5′-CANGANCGRAAYTGYTTRTT-3′ (complementary to amino acid sequence NKQFRSC conserved in SWS2s). Two oligonucleotide-pairs (SWSF/SWS1R and SWSF/SWS2R) were used in an attempt to amplify a fragment of coding sequences of SWS1 (∼170 bp) and SWS2 (∼360 bp), respectively, with single-strand cDNA from elephant shark eye as a template. The PCR cycles comprised an initial denaturation step at 95°C for 2 min; 35 cycles of 95°C for 30 sec, 50°C for 1 min, and 72°C for 1 min, followed by a final extension at 72°C for 5 min. Single-strand cDNA from Fugu eye was used as a control. The PCR products were sequenced to confirm their identity.
Elephant shark BAC for putative SWS1 locus
To search for an elephant shark ortholog of the SWS1 gene, we designed PCR oligonucleotides specific for the elephant shark Calu gene (syntenic with SWS1 in all vertebrates from teleosts to mammals) and screened a super-pooled DNA sample of an elephant shark Bacterial Artificial Chromosome (BAC) library (IMCB_Eshark BAC library cloned in pCCBAC-EcoRI; unpublished) and identified a BAC (#51A04) containing the Calu sequence. DNA from this BAC was sequenced to a depth of ∼4× coverage using the whole-genome shotgun sequencing approach and the sequences assembled with phred and phrap (Ewing et al. 1998).
Expression of visual pigment genes
Full-length cDNA for various genes were amplified as a single fragment by PCR using the 5′-RACE-ready cDNA as a template. Restriction sites for EcoRI and SalI were incorporated into sense and antisense oligonucleotide sequences, respectively. The sense oligonucleotide for each opsin contained a translation start codon (AUG) present within a Kozak consensus sequence to ensure efficient translation of the recombinant visual pigment. The antisense oligonucleotides were designed to replace the stop codon of the opsin protein with a SalI restriction site contained within a sequence that would encode a C-terminal bovine rod opsin 1D4 epitope (ETSQVAPA) that would be recognized by the anti-Rho1D4 monoclonal antibody. The PCR products were digested with EcoRI and SalI enzymes and cloned into the pMT4 expression vector and were sequenced completely to confirm that PCR had not introduced any mutations into the coding sequence (Franke et al. 1988).
The pMT4 vector containing each full-length opsin-coding sequence was transfected into human embryonic kidney (HEK-293T) cells by using a modified calcium phosphate (hydroxyapatite) coprecipitation method (Jordan et al. 1996) or GeneJuice Transfection Reagent (Novagen). In the hydroxyapatite transfection experiments, 300 μg of DNA and a N-(2-hydroxyethyl)-piperazine-N'-2-ethanesulfonic acid (HEPES) buffer containing 280 mM NaCl was used for each transfection. All other transfections were carried out using 210 μg of DNA and 630 μL of GeneJuice (Novagen). Twelve 140-mm plates were used per transfection; the cells were harvested 48 h post-transfection, and washed four times with 1× phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4) (pH 7.0). The visual pigments were regenerated in 1× PBS with 40 μM 11-cis retinal in the dark. 1% (w/v) dodecyl-maltoside and 20 mg/mL phenylmethanesulphonylfluoride (PMSF) was then added before passage over a CNBr-activated sepharose-binding column coupled to an anti-1D4 monoclonal antibody (Molday and MacKenzie 1983). In chloride-free experiments, all buffers used in the regeneration of elephant shark LWS1 and LWS2 visual pigments comprised of 1× PBS (pH 7.0), where 2.7 mM KCl and 137 mM NaCl were replaced with 2.7 mM KOH and 137 mM NaClO4, respectively, as according to Kleinschmidt and Harosi (1992).
Absorbance spectra were recorded in triplicate in the dark using a dual-path UV500 spectrophotometer (Spectronic Unicam). Pigments were bleached by exposure to light for 1 h before a second spectrum was recorded. The peak sensitivity value, the lambda max (λmax), for each pigment was determined by subtracting the bleached spectrum from the dark spectrum for each pigment to produce a difference spectrum. This was then fitted to a standard Govardovskii rhodopsin A1 template (Govardovskii et al. 2000) with a bleached retinal curve subtracted using an Excel spreadsheet to determine the λmax.
Phylogenetic analysis
Sequence alignments of elephant shark LWS1, LWS2, Rh2, and Rh1 opsin sequences, compared with LWS, SWS1, SWS2, Rh1, Rh2, RhA, and RhB nucleotide sequences across a variety of fishes (lampreys, elasmobranchs, and teleosts) and a Drosophila melanogaster (fruit fly) Rh4 (accession no. NM057353) outgroup, were generated by ClustalW (Higgins et al. 1996) and manually manipulated. Areas of disparity containing a high number of gaps, such as those near the 5′- and 3′-ends and other indels were removed, as they may introduce errors when estimating evolutionary distances. The resulting codon-matched alignments, which consisted of 909 nucleotides encoding for residues 16–321 (using the conventional numbering system of the bovine rod opsin polypeptide sequence), were subjected to phylogenetic analysis. Three different methods were used on the nucleotide alignment as follows. (1) A neighbor-joining tree (Saitou and Nei 1987), bootstrapped with 1000 replicates, was generated by applying a Kimura 2-parameter substitution matrix (Kimura 1980), a complete deletion, a homogenous pattern of nucleotide substitution among lineages, and a uniform rates of nucleotide substitution across all sites using the MEGA Version 3.1 computer package (Kumar et al. 2004). (2) Maximum likelihood was applied with a Kimura 2-parameter substitution matrix (Kimura 1980) and a gamma distribution parameter of 1 using PHYML (Guindon et al. 2005). The degree of support for internal branching was assessed by bootstrapping with 500 replicates. (3) A Bayesian probabilistic inference method was performed with a Metropolis Markov chain Monte Carlo (MCMC) algorithm using MrBayes 3.1.2 software (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). A general time-reversal (GTR) model (Lanave et al. 1984) was used, applying a gamma-distributed rate of variation across all sites with a proportion of invariable sites. Two simultaneous and independent runs starting from different random trees were performed for 850,000 generations with a chain sample frequency of 1000 generations. The first 25% (212) of all trees generated were discarded as burnin to allow for tree convergence and a low (close to 0.01) standard deviation of split frequencies. Resultant trees were visualized by Treeview 1.6.6 (Page 1996). Tajima's relative rate tests (Tajima 1993) were conducted using a χ2 distribution and one degree of freedom on two LWS nucleotide sequences, with the fruit fly Rh4 sequence as an outgroup, using the MEGA Version 3.1 computer package (Kumar et al. 2004). In this test, the null hypothesis assumes that the number of unique nucleotide substitutions along one lineage equals that of the sister lineage. A P- value of <0.05 was taken to indicate that the relative rates of evolutionary changes between two sequences were not equal and the null-hypothesis was rejected.
Acknowledgments
This work was supported in London by grants from the UK Biotechnology and Biological Research Council and the Leverhulme Trust, and in Singapore by the Biomedical Research Council of the A*STAR. We are grateful to Dr. Rosalie Crouch of the Storm Eye Institute, Medical University of South Carolina, USA for the provision of 11-cis-retinal, and Janine Danks, Justin Bell, and Terry Walker for their help in collecting elephant shark tissues.
Footnotes
-
↵3 Present address: Nuffield Laboratory of Ophthalmology, University of Oxford, John Radcliffe Hospital, Headley Way, Oxford, OX3 9DU, United Kingdom.
-
↵4 Corresponding authors.
↵E-mail d.hunt{at}ucl.ac.uk; fax 44-20-7608 6863.
↵E-mail mcbbv{at}imcb.a-star.edu.sg; fax 65-67791117.
-
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.084509.108.
-
- Received August 7, 2008.
- Accepted December 5, 2008.
- Copyright © 2009 by Cold Spring Harbor Laboratory Press