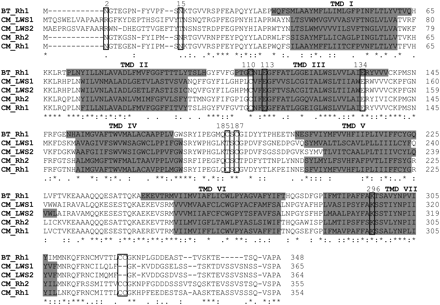
Alignment of the amino acid sequences of visual pigments expressed in the elephant shark (Callorhinchus milii; CM) and the common cow (Bos taurus, BT). (Rh1) Rod opsin; (LWS1, LWS2) long-wavelength-sensitive 1 and 2; (Rh2) cone rhodopsin 2 (middle-wavelength-sensitive). (*) An identical consensus residue between all four elephant shark opsin sequences and bovine Rh1. (: and .) A conserved or semiconservative amino substitution, respectively, with the codon-matched protein alignment. Gaps inserted to maintain a high degree of identity present between the opsin sequences derived from C. milii retina and bovine Rh1 are indicated by dashes (–). Seven putative transmembrane domains (TMDs) are indicated by gray shading. The TMDs shown for bovine rhodopsin were determined by crystallography (Palczewski et al. 2000). The putative positions of the TMDs for each elephant shark visual pigment were predicted online using TMHMM Server Version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Residues identified as being critical for correct opsin protein conformation (boxed). Comparison of the opsin amino acid sequences of C. milii and bovine rod opsin demonstrated that the critical residues involved in the maintenance of the tertiary structure of the opsin molecule are present. With the use of the conventional numbering system of the bovine rod opsin polypetide sequence, these key sites include (1) three conserved cysteine (C) residues at positions 110 (TMD3), 185 (ECD2), and 187 (ECD2) that are involved in disulphide bond formation (Karnik and Khorana 1990), except for a Thr (T) residue at position 185 in the elephant shark LWS1 and LWS2 opsins, which are also conserved throughout the rest of the vertebrate LWS opsin class; (2) a conserved glutamate (E) at position 113 (TMD3) that provides the negative counterion to the proton of the Schiff base (Sakmar et al. 1989); (3) a conserved glutamate (E) at position 134 (TM3) that provides a negative charge to stabilize the inactive opsin molecule (Cohen et al. 1993); (4) a conserved lysine (K) at position 296 (TM7) that is covalently linked to the chromophore via a Schiff base (Dratz and Hargrave 1983); (5) conservation of two cysteine (C) residues at putative palmitoylation positions 322 and 323 (Ovchinnikov et al. 1988) in both elephant shark Rh1 and Rh2 opsins, but not LWS1 and LWS2 opsins; (6) the presence of a number of Ser (S) and Thr (T) residues in the carboxyl terminus, which are potential targets for phosphorylation by rhodopsin kinases in the deactivation of metarhodopsin II (Palczewski et al. 1993; Sakmar and Fahmy 1996; Zhao et al. 1997); and (7) the conserved glycosylation sites at positions 2 and 15 (Sakmar and Fahmy 1996) in the MWS opsins (Rh1 and Rh2) identified in the retina of C. milii.











