Aberrant homeodomain–DNA cooperative dimerization underlies distinct developmental defects in two dominant CRX retinopathy models
- 1Molecular Genetics and Genomics Graduate Program, Division of Biology & Biomedical Sciences, Washington University in St. Louis, St. Louis, Missouri 63110, USA;
- 2Department of Ophthalmology and Visual Sciences, Washington University in St Louis, Saint Louis, Missouri 63110, USA;
- 3Department of Genetics, Washington University in St Louis, Saint Louis, Missouri 63110, USA;
- 4Department of Developmental Biology, Washington University in St Louis, Saint Louis, Missouri 63110, USA
Abstract
Paired-class homeodomain (HD) transcription factors (TFs) play essential roles in vertebrate development, and their mutations are linked to human diseases. One unique feature of a paired-class HD is cooperative dimerization on specific palindrome DNA sequences. Yet, the functional significance of HD cooperative dimerization in animal development and its dysregulation in diseases remains elusive. Using the retinal TF cone-rod homeobox (CRX) as a model, we have studied how blindness-causing mutations in the paired HD, p.E80A and p.K88N, alter CRX's cooperative dimerization, leading to gene misexpression and photoreceptor developmental deficits in dominant manners. CRXE80A maintains binding at monomeric WT CRX motifs but is deficient in cooperative binding at dimeric motifs. CRXE80A’s cooperativity defect impacts the exponential increase of photoreceptor gene expression in terminal differentiation and produces immature, nonfunctional photoreceptors in the CrxE80A retinas. CRXK88N is highly cooperative and localizes to ectopic genomic sites with strong enrichment of dimeric HD motifs. CRXK88N’s altered biochemical properties disrupt CRX's ability to direct dynamic chromatin remodeling during development to activate photoreceptor differentiation programs and silence progenitor programs. Our study provides in vitro and in vivo molecular evidence that paired-class HD cooperative dimerization regulates neuronal development and that dysregulation of cooperative binding contributes to severe dominant blinding retinopathies.
Homeodomain (HD) transcription factors (TFs) are essential for diverse biological processes in vertebrate development, including body plan specification, pattern formation, and cell fate specification (Mark et al. 1997; Hobert 2021; Leung et al. 2022). Paradoxically, for a protein domain that has evolved numerous functional specificities, it binds with high affinity to closely related DNA motifs that are typically only 5–6 bp long (Treisman et al. 1992; Wilson et al. 1996; Noyes et al. 2008; Bürglin and Affolter 2016). Thus, additional mechanisms are required to achieve the individual functions of homeoproteins (Wilson et al. 1993).
The paired-class HD family possesses a unique feature in that members of this class confer cooperative dimerization on specific dimeric DNA sequences. The “cooperative” interaction here is when the first HD–DNA half-site binding greatly enhances the binding of a second molecule to the other half-site (Fig. 1A). The paired-class HD cooperative dimerization solely relies on the 60-amino-acid HD, distinguishing it from the other HD families that require additional domains to form higher-order DNA-binding complexes (Hayashi and Scott 1990; Wilson et al. 1993). Yet, the functional importance of paired-class HD cooperative dimerization in development and its dysregulation in human diseases remains elusive.
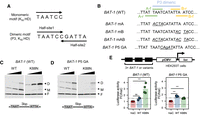
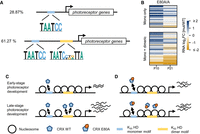
K88N mutation significantly increases CRX HD's cooperative binding and transactivation activity on BAT-1 sequence containing a P3 dimeric HD motif. (A) Diagrams depicting K50 HD preferred monomeric and dimeric P3 motifs. (B) Alignments showing WT BAT-1 probe sequences and variants. The P3 dimeric HD motif and four monomeric HD core motifs 5′-TAAT-3′ are labeled. (f) the core motif is on the forward strand; (r) the core motif is on the reverse strand. In BAT-1 variants, the mutated nucleotides are italicized and underlined. (C,D) EMSA gel images showing increasing amount of WT or K88N HD peptides bound to a fixed amount of BAT-1 (WT) or P5 GA control probes. The diagram, below each gel image shows the dimeric HD motif configuration and is labeled with the spacer length. (E) Schematics and barcharts of luciferase reporter assays comparing CRX WT and K88N transactivation activity at BAT-1 and variant enhancer sequences. Bars represent the mean. P-values of one-way ANOVA are annotated: (ns) >5 × 10−2, (***) ≤1 × 10−3.
We have studied CRX, a paired-class HD TF essential for photoreceptor cells in the retina, as a model to understand HD–DNA interactions in normal development and dominant blinding diseases (Tran and Chen 2014; Tran et al. 2014; Zheng et al. 2023; Zheng and Chen 2024). Photoreceptors are specialized neurons in the retina that sense light and initiate vision through the phototransduction process. In vertebrates, photoreceptors come in two major classes, rods and cones, that mediate vision in dim and bright light, respectively. Animal studies have demonstrated that Crx expression is activated in postmitotic photoreceptor precursors and is maintained throughout adult life (Chen et al. 1997; Furukawa et al. 1997; Muranishi et al. 2011). In the cellular context, disruption of CRX functions leads to significantly reduced enhancer activity of photoreceptor gene regulatory elements and the loss of gene expression, which ultimately results in degeneration of immature, nonfunctional photoreceptors (Furukawa et al. 1999; Roger et al. 2014; Tran et al. 2014; Ruzycki et al. 2015, 2018). Thus, CRX mainly functions as a positive regulator of photoreceptor development and functions in vivo. Coding sequence mutations in human CRX have been associated with at least three inherited retinal diseases (IRDs) that primarily affect photoreceptors: Leber congenital amaurosis 7 (LCA7; OMIM 613829), cone-rod dystrophy (CoRD) 2 (OMIM 120970), and retinitis pigmentosa (RP; OMIM 268000). CRX-associated retinopathies vary significantly in the ages of onset, severity, and disease progression (Tran and Chen 2014; Zheng and Chen 2024). The phenotype heterogeneity suggests that individual mutation may cause disease via distinct pathogenic mechanisms. Deciphering these mechanisms is, therefore, critical for informing the future development of therapeutic approaches.
CRX has two functional domains: the N-terminal DNA-binding domain (HD) and the C-terminal transcription effector domain. Disease-associated mutations are distributed across both domains, with amino acid substitutions primarily observed in the CRX HD (Tran and Chen 2014; Zheng and Chen 2024). To understand how HD mutations alter CRX–DNA interactions and lead to photoreceptor diseases, we have previously reported two human mutation knock-in mouse models (Zheng et al. 2023), each carrying a gain-of-function mutation, p.E80A (E80A) and p.K88N (K88N), which are associated with dominant LCA and dominant CoRD, respectively. Using an integrated approach that combines quantitative in vitro biochemical models, functional genomics, cellular profiling, and functional testing in mouse models, we found that E80A and K88N alter CRX DNA-binding specificity and produce distinct photoreceptor deficits in mutant mouse retinas. Yet, the proposed mechanisms were primarily based on analyzing CRX–DNA interactions at monomeric HD motifs both in vitro and in vivo.
Given that CRX is a paired-class HD TF, it is unclear whether E80A and K88N mutations affect CRX HD's cooperative dimerization and how mutant CRX activity interferes with WT CRX functions when both alleles are present, leading to the severe dominant photoreceptor deficits in developing mouse retinas. Here, we extend our multiomics approach to further elucidate the consequences of E80A and K88N mutations on CRX regulatory activities in developing photoreceptors. We will discuss how a single TF, CRX, through differential interactions with monomeric and dimeric DNA motifs, regulates different biological functions at different developmental stages and possibly in different cell types.
Results
CRX K88N but not WT or E80A HD confers strong cooperative dimerization on pRho BAT-1 probe
Paired-class HDs bind both monomeric and dimeric HD DNA motifs (Fig. 1A). Uniquely, paired-class HDs can cooperatively dimerize on specific dimeric motifs, historically known as P3 sequences (Fig. 1A, bottom; Wilson et al. 1993; Tucker and Wisdom 1999). In a P3 sequence, the two half-site core motifs 5′-TAAT-3′ are separated by a 3 bp spacer and form an approximate palindrome that places the two HDs in a head-to-head arrangement (Wilson et al. 1995). Recognition helix residues that determine paired-class HD's DNA-binding specificity at monomeric motifs also confer distinct cooperative dimerization properties, including the preferred spacer length and identity between the two 5′-TAAT-3′ core motifs and the magnitude of cooperativity. In the previous report, we found that disease-causing mutations E80A, K88N, and R90W, all located within the CRX HD recognition helix, differentially affect CRX HD DNA-binding specificity at monomeric sequences (Zheng et al. 2023). Here, we sought to understand whether any of the three mutations affect CRX HD's cooperative dimeric binding using electrophoretic mobility shift assays (EMSAs).
We chose the BAT-1 EMSA probe, an established model template to assay CRX HD's dimeric DNA binding (Fig. 1B; Chen et al. 1997, 2002). The BAT-1 sequence is a fragment in the promoter of rhodopsin, a gene that encodes the rod-specific photopigment and is a direct target of CRX in vivo. The BAT-1 fragment contains a central dimeric P3 sequence TAATCATATTA and additional overlapping monomeric HD motifs (Fig. 1B). We generated a series of BAT-1 variant fragments to explicitly interrogate the interactions between the two half-sites constituting the dimeric P3 motif (Fig. 1B). Specifically, in the BAT-1 P5 GA variant, the two half-sites are separated by a guanine (G) nucleotide, with each 6mer sequence preserved. The P5 configuration (5 bp spacer) has been shown to abolish cooperative dimerization of paired-class homeoproteins (Wilson et al. 1993) and thus was used as a control to visualize noncooperative dimeric binding events.
WT HD bound strongly to BAT-1 (WT) and P5 probes as monomeric and dimeric complexes but demonstrated no obvious cooperativity (Fig. 1C,D), as exemplified by the saturation of the monomeric band (M) before the gradual formation of the dimeric band (D). The difference in WT HD bound monomeric versus dimeric band intensities between P3 and P5 probes may be attributed to DNA shape features of the two half-sites that intrinsically affect HD–DNA interactions without explicitly changing the underlying sequences (Mathelier et al. 2016; Li et al. 2024). K88N HD showed strong dimeric binding with weak monomeric binding at the BAT-1 (WT) probe (Fig. 1C,D; Supplemental Fig. S1C), characteristic of cooperative dimerization in which the binding of one molecule stimulates the binding of a second molecule (Wilson et al. 1993; Tucker and Wisdom 1999). Increasing the spacer length (BAT-1 P5 GA) (Fig. 1D) or abolishing either half-site of the P3 sequence (BAT-1 mA and mB) (Supplemental Fig. S1C) resulted in diminished K88N HD dimeric binding, corroborating the essential P3 configuration and intact palindromic half-sites in mediating K88N HD's cooperative dimerization. The diminished K88N HD dimeric binding at the BAT-1 P5, mA, and mB probes unequivocally argues that K88N mutation does not render CRXK88N proteins obligatory dimers, and cooperative dimerization is mediated through specific HD–DNA interactions. In comparison, E80A HD bound stronger as monomeric complexes and weaker as dimeric complexes at both BAT-1 (WT) and P5 probes compared with WT HD (Supplemental Fig. S1A,B), which may inherently relate to E80A HD's reduced binding specificity at monomeric sequences (Zheng et al. 2023); R90W HD bound poorly to either BAT-1 (WT) or P5 probe, consistent with R90W being a loss-of-function mutation that reduces CRX HD's overall DNA-binding affinity (Chen et al. 2002). In summary, only CRX K88N but not WT or E80A HD confers strong cooperative dimerization on the BAT-1 probe containing a P3 dimeric HD motif.
CRXK88N cooperative dimerization mediates strong reporter gene activation in HEK293T cells
To determine the consequences of K88N HD's enhanced cooperative dimerization on CRX's transactivation activity, we performed luciferase reporter assays in the HEK293T cells with enhancers harboring three tandem repeats of BAT-1 or variant sequences identical to that used in EMSAs. Consistent with EMSA results, CRXK88N demonstrated strong activator activity at a 3 × BAT-1 WT enhancer harboring the intact P3 dimeric sequence but much weaker activity at all BAT-1 variants (Fig. 1E; Supplemental Fig. S1D). Thus, CRXK88N is a competent transcription activator, and CRXK88N’s cooperative dimerization on BAT-1 P3 sequence mediates strong gene activation, likely through stabilizing the dimeric binding complexes.
CRXWT only significantly activated the 3 × BAT-1 P5 and mA enhancers but not the 3 × BAT-1 WT and mB enhancers (Fig. 1E; Supplemental Fig. S1D). In the BAT-1 mA sequence, the suboptimal 5′-TAATCA-3′(A-f) is destroyed, and a single WT CRX consensus monomeric motif 5′-TAATCC-3′ (B-f) remains intact. Because typical B-form DNA is arranged in helical turns of 10.5 bp and paired-class HD's cooperative dimerization involves DNA conformational changes (Wilson et al. 1995), the orientation of CRX protein binding relative to the transcription start site and the relative orientation of two CRX proteins in the dimeric binding complex are different in different BAT-1 variants. These orientation differences may alter the interaction surfaces for additional factors that bind to CRX and ultimately lead to the creation of the transcription initiation complex.
K88N enhances but E80A impairs CRX HD's cooperative dimerization at various P3 sequences in vitro
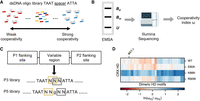
Although WT CRX HD did not show apparent cooperative dimerization at the BAT-1 (WT) probe, it is unclear whether it can form cooperative dimers on other P3 sequences. To characterize CRX HD's DNA-binding cooperativity unbiasedly, we adapted a high-throughput in vitro assay, Coop-seq, that determines protein–DNA-binding cooperativity by sequencing (Fig. 2A,B; Chang et al. 2017; Hu et al. 2017). Coop-seq was developed based on traditional EMSAs, which allow the physical separation of distinct binding complexes. Coop-seq enables us to accurately measure the cooperativity parameters—interactions between two HD half-sites—for a library of dimeric HD DNA motifs in parallel and quantitatively compare the cooperativity of different CRX HDs. Based on HD–DNA interaction models and previous Spec-seq-generated monomeric CRX HD DNA-binding models (Zheng et al. 2023), we designed a Coop-seq library containing all possible dimeric P3 spacer sequences TAATNNNATTA (Fig. 2C).
E80A and K88N mutations differently affect CRX HD DNA-binding cooperativity at P3 sequences. (A,B) Schematics showing the Coop-seq experimental pipelines. dsDNA oligo pools of P3 and/or P5 Coop-seq library are incubated with different HD peptides. The dimeric and monomeric binding complexes are separated from unbound DNAs by EMSA. DNAs are extracted from all three DNA bands and subjected to quantification by Illumina sequencing. (Bd) Dimeric band, (Bm) monomeric band, and (U) unbound band. (C) Diagram depicting the Coop-seq library design and strategy to match a P3 sequence with a P5 counterpart. Exact oligo sequences can be found in Supplemental Table S1. (D) Heatmap comparing the relative cooperativity of WT and variant CRX HDs on P3 and P5 libraries (ωp3/ωp5). Note the relative cooperativity is presented in the logarithmic scale and ordered by unsupervised hierarchical clustering. The ordered relative cooperativity matrix can be found in Supplemental Table S3.
As a control, we first tested a P5 library (TAATNNGNNATTA) (Fig. 2C), which is expected to limit cooperativity between half-sites (Fig. 1D; Supplemental Fig. S1B; Wilson et al. 1993; Tucker and Wisdom 1999). We obtained the DNA-binding cooperativity index ω of WT and mutant HDs at the P5 library using bacterially expressed and affinity-purified HD peptides as previously described (Methods). As expected, WT and mutant CRX HDs showed weak cooperativity at P5 sequences (Supplemental Fig. S2A–D; Supplemental Table S2). It supports the idea that CRX HDs primarily bind nonoverlapping half-sites independently, consistent with homeoproteins’ high-affinity binding at monomeric HD motifs.
Next, we compared CRX HDs’ cooperativity profiles on the preferred P3 configuration sequences. To visualize the specific cooperative interactions elicited by the P3 configuration, we normalized the cooperativity index ω at P3 sequences (ωp3) by their P5 counterparts (ωp5; Methods; Fig. 2C). WT HD showed moderate cooperativity at a small subset of P3 sequences (Fig. 2D). Close examination of the P3 sequence spacers of this subset revealed an enrichment of the guanine (G) and cytosine (C) bases (Supplemental Table S3), which are known to be preferred by the Lys50 (K50) HD subfamily of paired-class homeoproteins, including CRX. R90W HD showed a similar cooperativity profile as WT HD with slightly increased cooperativity at a few P3 sequences. This is consistent with R90W being a loss-of-function mutation that reduces the overall HD–DNA-binding affinity without selectively altering the specific HD–DNA contacts (Chen et al. 2002; Zheng et al. 2023). E80A HD exhibited reduced cooperativity compared with WT HD, whereas K88N HD showed enhanced cooperativity at all P3 sequences. A previous study found that a Gln50 (Q50) paired-class HD shows more than 10-fold stronger cooperativity than a K50 HD (Wilson et al. 1993). Because K88N mutation alters CRX HD's DNA-binding specificity at monomeric motifs to one that mimics a natural Q50 HD (Zheng et al. 2023), K88N HD's enhanced cooperativity at dimeric motifs can be attributed to its similarity to a Q50 HD.
Collectively, in vitro protein–DNA binding results indicate that E80A mutation impairs CRX HD's DNA-binding cooperativity at specific dimeric HD motifs, whereas K88N mutation drastically alters both specificity and cooperativity. The luciferase reporter assays highlight the functional distinctions between cooperative dimeric binding at P3 sequences in contrast to noncooperative dimeric binding and individual binding to monomeric sites.
CrxK88N/+ and CrxK88N/N retinas show severely decreased accessibility at CREs enriched for K50 HD motifs
Next, we sought to understand the roles of CRX's monomeric and cooperative dimeric binding on the regulation of photoreceptor development using CrxE80A and CrxK88N mouse models established in our previous study (Zheng et al. 2023). We asked how E80A and K88N mutations affect CRX's ability to facilitate chromatin remodeling by performing retinal ATAC-seq on postnatal day (P) 14 WT, heterozygous, and homozygous mutant retinas. For concision, we use CrxE80A and CrxK88N when both heterozygous and homozygous mutants are discussed. The ATAC-seq results were analyzed in conjunction with our published P14 CRX ChIP-seq data. We asked how E80A and K88N mutation-specific changes in HD–DNA interactions affect the chromatin landscape in individual mutant models and how perturbed epigenome relates to photoreceptor gene misexpression.
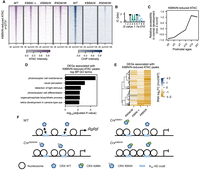
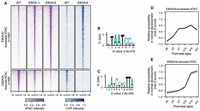
Because CRXWT and CRXK88N prefer different monomeric HD motifs in vitro and in vivo (Zheng et al. 2023), we predicted that the CrxK88N/N retinas show similar chromatin remodeling defects as the loss-of-function CrxR90W/W model and that CRXWT proteins in the CrxK88N/+ retinas can bind canonical CRX binding sites and facilitate chromatin remodeling. We found that the CrxK88N/N retinas showed more significant chromatin accessibility loss at canonical CRX binding sites than the loss-of-function CrxR90W/W retinas and that the CrxK88N/+ retinas showed reduced accessibility similar to that of the CrxR90W/W retinas (Fig. 3A). Genomic regions that showed defective chromatin remodeling in the CrxK88N/+ and CrxK88N/N retinas were enriched for the K50 HD motifs (Fig. 3B), gain accessibility in normal postnatal retinal development (Fig. 3C), and regulate genes important for photoreceptor structures, functions, and maintenance (Fig. 3D). The impaired chromatin remodeling at canonical CRX binding sites led to significant photoreceptor gene downregulation in the developing CrxK88N retinas (Fig. 3E). Thus, defects in chromatin remodeling at regions with K50 HD motifs underlie defective photoreceptor differentiation in young adults in CrxK88N/+ and CrxK88N/N mice (Fig. 3F; Zheng et al. 2023).
CrxK88N/+ retinas show defective chromatin remodeling at photoreceptor CREs enriched with K50 HD motifs. (A) Heatmaps depicting the normalized ATAC-seq or CRX ChIP-seq signal intensities at CrxK88N-reduced accessible ATAC-seq peaks. (B) PWM logo and enrichment significance E-value of the STREME de novo discovered HD motif. (C) Line plot showing the average developmental accessibility kinetics of CrxK88N-reduced ATAC-seq peaks. The developmental ATAC-seq data are from Aldiri et al. (2017). (D) Barchart showing biological process (BP) Gene Ontology (GO) term enrichment of differentially expressed genes adjacent to CrxK88N-reduced ATAC-seq peaks. (E) Heatmap comparing the P10 expression changes of CrxK88N-reduced ATAC-seq peaks associated genes in different Crx mutant retinas. The gene set is identical to that in D. (F) Schematics depicting chromatin remodeling defects at photoreceptor regulatory regions in the CrxK88N/+, CrxK88N/N, and CrxR90W/W retinas.
CrxK88N/+ and CrxK88N/N retinas show increased accessibility at CREs enriched for Q50 HD motifs
Because the CrxR90W/+ retinas, expressing a single dose of CRXWT proteins, show largely WT phenotypes (Tran et al. 2014), the severe chromatin remodeling defects and photoreceptor gene misexpression in the CrxK88N retinas are likely attributed to CRXK88N ectopic activities. The CrxK88N-increased accessibility ATAC-seq peaks coincided with ectopic CRXK88N binding but showed comparably low accessibility in the CrxR90W/W and WT retinas (Fig. 4A). De novo motif searching on sequences under CrxK88N-increased ATAC-seq peaks revealed both dimeric and monomeric Q50 HD motifs (Fig. 4B), consistent with in vitro found alterations in K88N HD–DNA-binding specificity and cooperativity. The CrxK88N retina ectopically enriched dimeric HD motifs exhibit no base preferences within the 3 bp spacer (Fig. 4B, positions 5–7), in line with K88N HD's strong cooperativity at nearly all P3 Coop-seq library sequences (Fig. 2D). Collectively, these observations suggest that ectopic CRXK88N activity at Q50 HD motifs rather than loss of CRXWT activity mediates chromatin accessibility increase at ectopic sites in the CrxK88N/+ and CrxK88N/N retinas.
CRX K88N ectopic activity at Q50 HD motifs impedes the silencing of progenitor regulatory programs in developing photoreceptors. (A) Heatmaps depicting the normalized ATAC-seq or CRX ChIP-seq signal intensities at CrxK88N-increased accessible ATAC-seq peaks. (B) PWM logo and enrichment significance E-value of STREME de novo discovered HD motifs. (C) Line plot showing the average developmental accessibility kinetics of CrxK88N-increased ATAC-seq peaks. The developmental ATAC-seq data are from Aldiri et al. (2017). (D) Heatmap depicting the log odds ratio enrichment of embryonic day (e) 14.5 or adult retinal VSX2 binding sites under CrxK88N-increased ATAC-seq peaks. ⋂ indicates the intersection of VSX2 ChIP and K88N/N up ATAC peaks. P-values of Fisher's exact tests are indicated. The VSX2 ChIP-seq data are from Bian et al. (2022). (E) PWM logo and significance E-value of STREME de novo discovered basic helix–loop–helix (bHLH) motif under CrxK88N-increased ATAC-seq peaks. PWM logos of selected retinal progenitor/neurogenic bHLH TFs are given for comparison. JASPAR IDs of the selected TFs can be found in Methods. (F) Barchart showing BP GO term enrichment of genes adjacent to CrxK88N-reduced ATAC-seq peaks.
CrxK88N-increased accessibility CREs show progenitor cell regulatory signatures and are developmentally silenced during photoreceptor differentiation
Last, we sought to understand the functional significance of CRXK88N-associated chromatin accessibility increase. Different from previously characterized mutant CRX proteins that recognize K50 HD motifs, CRXK88N prefers Q50 HD motifs that are recognized by Q50 HD TFs highly expressed in retinal progenitor cells (Bassett and Wallace 2012; Zagozewski et al. 2014). In the developing mouse retinas, these Q50 HD TFs coordinate the dynamic chromatin landscape changes during retinal neurogenesis and are downregulated in differentiating photoreceptors (Zibetti et al. 2019; Bian et al. 2022). We predicted that CRXK88N ectopic activities affect the chromatin landscape changes at progenitor CREs that are usually bound by progenitor Q50 homeoproteins.
Using a previously published ATAC-seq data set of normal mouse retinal development (Aldiri et al. 2017), we found that CrxK88N-increased ATAC-seq peaks showed the strongest accessibility at neonatal ages P0 and P3, followed by a gradual decrease in accessibility as photoreceptors undergo differentiation (Fig. 4C). Reanalysis of a published VSX2 (Q50 HD TF) retinal ChIP-seq data (Bian et al. 2022) showed that embryonic day (E) 14.5 but not adult VSX2 binding is enriched at the CrxK88N-increased ATAC-seq peaks (Fig. 4D). In the embryonic retina, VSX2 is expressed in retinal progenitor cells; in the adult retina, VSX2 expression is maintained in bipolar cells and Müller glia but is lost in mature photoreceptors (Liu et al. 1994; Burmeister et al. 1996; Rowan and Cepko 2004). Thus, the enrichment of embryonic VSX2 binding and the depletion of adult VSX2 binding at CrxK88N-increased ATAC-seq peaks suggests that CRXK88N ectopic activity is likely impeding the silencing of progenitor chromatin states instead of directing photoreceptor precursors into alternative cell lineage programs. Additionally, de novo motif enrichment analysis of sequences under CrxK88N-increased ATAC-seq peaks found patterns characteristic of basic helix–loop–helix (bHLH) neurogenic TF consensuses (Fig. 4E), corroborating the potential functionality of CrxK88N-increased ATAC-seq peaks in regulating neurogenic programs during normal development. Gene Ontology (GO) analysis revealed that genes adjacent to CrxK88N-increased ATAC-seq peaks were implicated in general neuronal development (Fig. 4F). Collectively, these pieces of evidence suggest that instead of initiating accessibility de novo, CRXK88N’s ectopic activities at Q50 HD motifs maintain the accessibility of chromatin regions that are developmentally closed, which may create a chromatin environment that is inhibitive to TFs that regulate photoreceptor differentiation.
CrxE80A retinas show opposite chromatin accessibility changes at CREs enriched for monomeric and dimeric K50 HD motifs
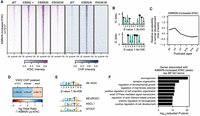
Different from the K88N mutation that drastically alters CRX HD's DNA-binding specificity, the E80A mutation does not affect CRX HD's DNA sequence preference per se but reduces binding specificity at monomeric motifs (Zheng et al. 2023) and impairs binding cooperativity at specific dimeric motifs (Fig. 2C). We asked if these changes differentially affected CRXE80A’s ability to modulate chromatin remodeling. Among the consensus ATAC-seq peaks identified in WT, CrxE80A/+, and CrxE80A/A retinas, a set of peaks (n = 1681) showed significantly increased ATAC-seq signal intensity in the CrxE80A/A retinas compared with WT, with the CrxE80A/+ retinas showing intermediate intensity at these loci (Fig. 5A). The CrxE80A-increased ATAC-seq peaks showed increased CRXE80A binding in the CrxE80A/A retinas and were enriched for the WT CRX consensus monomeric K50 HD motifs (Fig. 5B). No additional TF motif was found significantly enriched in these peaks, suggesting a major contribution from CRXE80A binding and activity. A smaller subset of peaks (n = 485) showed significantly decreased ATAC-seq signals in the CrxE80A/A retinas compared with WT, again with the CrxE80A/+ retinas showing intermediate signal reduction (Fig. 5A). The CrxE80A-reduced ATAC-seq peaks showed diminished CRXE80A binding in the CrxE80A/A retinas correlating with defective chromatin remodeling. De novo motif searching of sequences under the CrxE80A-reduced ATAC-seq peaks identified a dimeric P3 sequence pattern resembling dimeric K50 HD motifs, highlighted by a preference for cytosine (C) at the spacer positions 5 and 6 (Fig. 5C).
CrxE80A retinas show defective chromatin remodeling at CREs enriched for dimeric K50 HD motifs. (A) Heatmaps depicting the normalized ATAC-seq or CRX ChIP-seq signal intensities at CrxE80A differentially accessible ATAC-seq peaks. (B,C) PWM logos and enrichment significance E-values of STREME de novo discovered HD motifs under CrxE80A differentially accessible ATAC-seq peaks. (D,E) Line plots showing the average developmental accessibility kinetics of CrxE80A differentially accessible ATAC-seq peaks. The developmental ATAC-seq data are from Aldiri et al. (2017).
Because CRX proteins can, theoretically, bind noncooperatively to the two half-sites constituting a P3 dimeric motif as observed in EMSAs with BAT-1 probes (Fig. 1B), we sought to determine what sequence features of the dimeric motifs under CrxE80A-reduced ATAC-seq peaks rendered them susceptible to defects in CRX's cooperative dimerization. We first identified instances of dimeric K50 HD motifs under CrxE80A-reduced ATAC-seq peaks by scanning the DNA sequences with FIMO (Grant et al. 2011) using the dimeric K50 HD motif position weight matrix (PWM) in Figure 5C at a P-value threshold of 1 × 10−3 (Methods). We then estimated how well WT CRX proteins can bind individually to the two half-sites by calculating their relative binding affinities using a CRX monomeric binding PWM model (Lee et al. 2010). We found that these dimeric motifs often consist with two low-affinity half-sites, suggesting cooperative dimerization may be crucial to facilitate CRX's stable binding (Supplemental Fig. S3A). This is distinct from previous analyses that have focused mostly on the highest-affinity (not the strongest cooperativity) dimeric K50 HD motifs on nonchromatin templates (Wilson et al. 1993; Hughes et al. 2018). Our observations suggest that cooperative dimerization is critical for CRX regulatory activities at specific dimeric K50 motifs in vivo and in the chromatin context. Impaired cooperative dimerization likely reduces CRXE80A’s stable binding at these dimeric K50 HD motifs and, in turn, affects the chromatin remodeling activity.
CrxE80A differentially accessible CREs exhibit distinct chromatin remodeling kinetics in normal development
Previously, we observed that photoreceptor differentiation is perturbed in a cell-type-specific and developmental stage–specific manner in CrxE80A retinas (Zheng et al. 2023). Rod photoreceptors show precocious early differentiation but defective terminal differentiation, whereas cone photoreceptors show a lack of differentiation at both stages. We asked whether these patterns were associated with CRXE80A’s differential impacts on genomic regions of different regulatory functions. It is important to note that the mouse retina is rod-dominant; thus, global patterns in bulk ATAC-seq signals mainly reflect chromatin accessibility landscape in rods, and regulatory elements for cones need to be evaluated in a gene-specific manner. Using a previously published ATAC-seq data set of normal mouse retinal development (Aldiri et al. 2017), we found that both CrxE80A-increased and CrxE80A-reduced ATAC-seq peaks gain accessibility during postnatal retinal development (Fig. 5D,E). Yet, the two sets of peaks differ in their kinetics of accessibility gain. In WT retinas, the CrxE80A-increased ATAC-seq peaks exhibit a strong increase in accessibility during early photoreceptor development, from P0 to P7, followed by a moderate change from P7 to P21. The CrxE80A-reduced ATAC-seq peaks are characterized by an exponential gain in accessibility between P10 and P14 with smaller changes before P10 and after P14. The P10–P14 time points represent a critical window of photoreceptor differentiation characterized by a significant change in the photoreceptor transcriptome (Kim et al. 2016) and the elaboration of outer segments (OSs), the subcellular structures in which phototransduction occurs (Swaroop et al. 2010). The distinct developmental accessibility kinetics suggest that CRXE80A activity at monomeric and dimeric K50 HD motifs might affect gene expression at different stages of photoreceptor development in the mutant mouse retinas.
Monomeric and dimeric K50 HD motifs associated with stage-specific photoreceptor gene misexpression in the CrxE80A retinas
To identify the likely direct impacts of CRXE80A mutant activity on gene expression, we focused on the “CRX-dependent activated genes” (CRX-DAGs) defined previously (Zheng et al. 2023). CRX-DAGs are adjacent to at least one CRX binding site, and their expressions are significantly reduced in the loss-of-function model CrxR90W/W. CRE–gene association analysis using ATAC-seq-identified CREs that also overlap with CRX binding revealed that many CRX-DAGs are potentially under combinational regulations of monomeric and dimeric K50 HD motifs (Methods; Supplemental Table S6). Specifically, 28.87% of CRX-DAGs are associated with CREs that only contain monomeric K50 HD motifs, whereas 61.27% of CRX-DAGs are associated with one or more CREs that contain both monomeric and dimeric K50 HD motifs (Fig. 6A). By analyzing a previously published RNA-seq data set of normal mouse retinal development (Aldiri et al. 2017), we found that genes regulated by both monomeric and dimeric K50 HD motifs display a broader range of expression changes with genes critical for rod photoreceptor terminal differentiation displaying an exponential increase, such as Esrrb, Gnat1, and Rho (Supplemental Fig. S4A,B). This suggests that CRX's action on dimeric motifs may be associated with unique gene expression changes for a specific subset of target genes.
CRX E80A differential activity at monomeric and dimeric K50 HD motifs contributes to gene misexpression at different stages of photoreceptor development. (A) Diagrams of photoreceptor genes regulated solely by monomeric K50 HD motifs (top) or combinationally by monomeric and dimeric K50 HD motifs (bottom). For simplicity, representative motif logos are shown. The relative position of the motifs is arbitrary. (B) Heatmap comparing the CRX-DAG expression changes in the CrxE80A/A retinas at ages of postnatal day (P) 10 and P21. The gene sets in the heatmaps are as defined in A. (C,D) Schematics demonstrating the K50 HD division-of-labor model in regulating photoreceptor epigenome and transcriptome at different stages of development.
To understand how altered CRXE80A–DNA interactions at monomeric and dimeric K50 HD motifs relate to photoreceptor gene misexpression in the developing (P10) and mature (P21) CrxE80A mouse retinas, we reanalyzed the RNA-seq data generated in our previous study (Zheng et al. 2023). In the P10 CrxE80A/A retinas, CRX-DAGs associated either solely with monomeric K50 HD motifs or also with dimeric K50 HD motifs showed overexpression compared with the WT (Fig. 6B, P10 columns). Because regulatory elements enriched for the monomeric K50 HD motifs display an early chromatin remodeling profile in normal development (Fig. 5D), it is likely that CRXE80A promiscuous binding at monomeric K50 HD motifs (Zheng et al. 2023) accelerated the chromatin remodeling and/or directly enhanced the expression of CRX-DAGs in the developing CrxE80A/A mutant retinas. In the P21 adult CrxE80A/A retinas, expression of the P10 CrxE80A/A-overexpressed genes showed two patterns: Genes associated solely with monomeric K50 HD motifs became comparable to WT or remained overexpressed but of a much lower magnitude; genes associated with both monomeric and dimeric K50 HD motifs became comparable to the WT and even significantly downregulated (Fig. 6B, P21 columns). The CrxE80A/+ retinas showed similar patterns of gene misexpression but at a less severe degree (Supplemental Fig. S5A). The selective downregulation of dimeric K50 HD motif–associated genes may be explained by CRXE80A’s impaired cooperative dimerization and subsequently defective chromatin remodeling at regulatory elements enriched for the dimeric HD motifs. Genes encoding structural proteins of photoreceptor OSs and molecules involved in the second messenger cascade of the visual cycle are enriched in this specific set of CRX target genes (Supplemental Fig. S5B; Supplemental Table S6). The precise regulation of these genes is fundamental to the integrity of photoreceptor OSs and functions (Purves and Williams 2001). Perturbations of these genes have been associated with inherited retinal dystrophies that affect rods, cones, or both and cause blindness (García Bohórquez et al. 2021). Thus, the underexpression of these genes likely underlies the defective photoreceptor terminal differentiation and functions in the CrxE80A retinas.
There is a subset of dimeric motif–associated CRX-DAGs that were downregulated at P10 and become more severely down at P21 (Fig. 6B; Supplemental Fig. S5A). This gene set is implicated in cone photoreceptor structures and functions. In the mouse retinas, cones are born embryonically whereas most rods are born postnatally, and their differentiation progresses differently. At the postnatal ages examined, it is likely that we captured different stages of cone and rod photoreceptor differentiation.
Monomeric and dimeric K50 HD motifs demonstrate regulatory activity changes in explant retinas
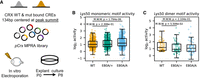
The analyses above clearly demonstrate a close relationship between dysregulated CRXE80A activity at different K50 HD motifs and gene misexpression in the CrxE80A retina. However, it remains unclear whether CRXE80A acts directly or through modulating chromatin accessibility to affect gene expression. To evaluate the relative contribution of CRXE80A–DNA interactions to transcription regulation of nonchromatin templates, we selected and tested a representative set of CRX-bound cis-regulatory elements (CREs) in episomal plasmids with massively parallel reporter assays (MPRAs) in explant WT and mutant mouse retinas (Methods; Fig. 7A; Supplemental Fig. S7A,B). We measured the regulatory activities of genomic CREs with intact HD motifs and with activity-abolishing mutations in the HD motifs. The regulatory activity difference between each pair of CREs with intact and mutated HD motifs yields an HD motif activity measurement that reflects the direct functional consequences of altered CRX–HD motif interactions (Methods). Comparison between genotypes found a significant increase in monomeric K50 HD motif activity in the CrxE80A/A retinas (Fig. 7B), suggesting that CRXE80A binding at monomeric motifs can directly drive increased target gene expression in the developing retinas. Different from the monomeric motifs, dimeric K50 HD motifs showed similar degree of activity reduction in both CrxE80A/+ and CrxE80A/A retinas (Fig. 7C). Because K50 HD's dimeric binding is of much lower affinity compared with its monomeric binding (Wilson et al. 1993), it is possible that proteins generated from the single allele of CrxWT is insufficient to produce stable binding at dimeric K50 HD motifs in the CrxE80A/+ retinas. The MPRA results, combined with alterations of photoreceptor epigenome in vivo, suggest that CRXE80A misregulates gene expression by acting through both chromatin remodeling and direct transactivation pathways.
Monomeric and dimeric K50 HD motifs show different regulatory activity changes in retinal explant MPRAs. (A) Schematics showing explant retinal MPRA experimental pipeline. (B,C) Box and strip plots comparing monomeric (B) or dimeric (C) K50 HD motif activities in explant cultured WT, CrxE80A/+, and CrxE80A/A retinas. (B) CREs overlapped with ATAC-seq peaks that were not significantly reduced in the CrxE80A retinas are plotted. (C) CREs overlapped with ATAC-seq peaks that were significantly reduced in the CrxE80A retinas are plotted. P-values of Mann–Whitney–Wilcoxon tests are annotated.
Discussion
HD TFs control the development and functions of many tissues. The retina is an excellent system for understanding how a single HD TF achieves functional specification in different cell types and at different times in development. The development and homeostasis of retinal photoreceptor cells are controlled by a master HD TF, CRX. Human mutations in CRX cause a spectrum of IRDs that show significant heterogeneity in clinical phenotypes.
Because CRX is expressed in both cone and rod photoreceptors, both during development and in adults, how does one explain the differential consequences of CRX mutations? In this study, we have extended our previous investigations on two CRX HD missense mutations, E80A and K88N, that are associated with dominant CoRD and dominant LCA in humans. Besides the previously identified differential impacts on DNA-binding specificity at monomeric motifs (Zheng et al. 2023), E80A and K88N mutations also differently alter CRX HD's cooperative binding at dimeric HD sequences. The mutation-specific effects on DNA-binding specificity and cooperativity underlie the chromatin landscape changes that explain the distinct dominant photoreceptor gene misexpression patterns in the mutation knock-in mouse retinas. Unlike other Crx mouse models, no obvious photoreceptor degeneration was observed in either CrxE80A or CrxK88N retinas at the ages examined (Zheng et al. 2023), suggesting that the defects in chromatin remodeling and gene expression are largely attributed to changes in CRX's intrinsic regulatory functions instead of changes in cell number. Analysis of the epigenome and transcriptome dynamics in normal development and in the CrxE80A and CrxK88N mutant retinas highlights an underappreciated role of DNA-mediated CRX cooperative dimerization in ensuring proper temporal chromatin and gene expression changes in photoreceptor development.
HD residue 50 (corresponding to CRX K88) determines paired-class HDs’ DNA-binding specificity at monomeric and dimeric HD motifs as well as HD's cooperative binding at palindrome dimeric motifs (Hanes and Brent 1989, 1991; Treisman et al. 1989; Wilson et al. 1993). In addition to the difference in sequence preference at their respective monomeric consensus motifs, a K50 HD bound with an order of magnitude higher affinity and more than 100-fold in the half-life of the binding complex than a Q50 HD (Ades and Sauer 1994); at dimeric consensus motifs, a Q50 HD bound with 10-fold stronger cooperativity than a K50 HD (Wilson et al. 1993). Asparagine (N) is structurally similar to glutamine (Q) in that both of them contain amide (NH2) groups in their respective side chains, and they differ only by one methylene group. Expectedly, the K88N mutation alters CRX HD's DNA-binding specificity from K50 to that resembling a Q50 HD (Zheng et al. 2023) and enhances cooperative dimerization both at the BAT-1 P3 probe (Fig. 1C) and at Coop-seq P3 oligos (Fig. 2D). The drastic difference between WT and K88N HDs in the scale and spectrum of cooperativity agrees with the random-site selection assays (Wilson et al. 1993) in which a Q50 HD bound strongly to many P3 sequences, yielding a 5′-TAATPyNPuATTA-3′ consensus, whereas a K50 HD only selected the monomeric consensus 5′-TAATCC-3′ under the same conditions as the Q50 HD, and three additional rounds of selection were required to recover the dimeric consensus 5′-TAATCCGATTA-3′. In sum, CRX K88N HD resembles a natural Q50 HD in both monomeric and cooperative dimeric binding.
In the developing WT mouse retinas, CRX binding at K50 HD motifs is essential for photoreceptor differentiation by facilitating chromatin remodeling and regulating target gene expression (Ruzycki et al. 2018). CRXK88N’s diminished activity at canonical CRX motifs leads to defective chromatin remodeling at photoreceptor CREs and loss of target gene expression in the CrxK88N/N retinas (Fig. 3; Zheng et al. 2023). Yet, K88N mutation's dominant inheritance pattern and the more severe gene misexpression in the CrxK88N/N retinas than the loss-of-function CrxR90W/W retinas (Fig. 3E) suggests that CRXK88N’s ectopic activities at Q50 HD motifs have a significant impact on photoreceptor development. Q50 HD motifs are recognized by many HD-containing TFs expressed in the retinal progenitor cells (Bassett and Wallace 2012; Heavner and Pevny 2012; Zagozewski et al. 2014). These progenitor Q50 HD TFs promote retinal progenitor cell proliferation and thus are inhibitory to neurogenesis and subsequently postmitotic differentiation (Burmeister et al. 1996; Green et al. 2003; Gordon et al. 2013). In CrxK88N retinas, CRXK88N may function as an ectopic Q50 HD TF in postmitotic photoreceptors to maintain the cells at an undifferentiated state. Mechanistically, it remains unclear what the molecular pathways are that respond to CRXK88N’s ectopic activities and in turn modulate the epigenetic status of CREs crucial for photoreceptor development. Differentially expressed genes near CRXK88N’s ectopic accessible regions do not show significant enrichment in a single GO or pathway. This suggests the epigenomic and transcriptomic changes in CrxK88N retinas may be a collective consequence of perturbations on many biological processes. In addition, data in our study were generated from whole-retina samples, which can mask small cell-type-specific changes, and the time points investigated in this study do not capture photoreceptor precursors, which may be the critical stage of CRXK88N’s actions. Single-cell profiling or molecular characterization of purified photoreceptors from WT and CrxK88N mutant retinas at different ages will shed light on the antagonistic interplays between CRXK88N and CRXWT functions. These targeted analyses of pure cell populations will be crucial to understanding whether the developmental accessibility pattern of CrxK88N-ectopic regions in Figure 4C was caused by changes in cell-type proportion or specific changes in postmitotic photoreceptors. In summary, CRX target specificity is not only critical for activating photoreceptor developmental programs but also crucial for the proper silencing of early programs that could be inhibitory to photoreceptor differentiation at later stages.
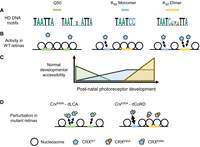
Different from K88N's global impacts on CRX HD–DNA interactions, E80A mutation increases HD's promiscuous binding at monomeric K50 HD motifs (Zheng et al. 2023) but reduces cooperative binding at dimeric K50 HD motifs (Fig. 2D). CRXE80A’s differential interactions with K50 HD motif subtypes parallel the hyperactivation of rod photoreceptor genes in developing CrxE80A retinas and the hypoactivation of the same set of genes in adult CrxE80A retinas (Zheng et al. 2023). These observations suggest a K50 HD motif division-of-labor model in which early-stage photoreceptor development is mediated by CRX interactions with monomeric K50 HD motifs, whereas terminal differentiation additionally relies on interactions with dimeric K50 HD motifs that specifically require cooperative dimerization (Fig. 8). This dichotomy of K50 HD motif usage likely relates to the different CRX–chromatin interaction kinetics collectively determined by the underlying DNA sequences and CRX concentrations (Fig. 5D,E; Supplemental Fig. S4C,D). Specifically, CRX binds monomeric K50 HD consensus motifs with high affinity, which can activate gene expression at relatively low CRX concentration, such as in photoreceptor precursors. However, CRX binding easily saturates at high-affinity monomeric motifs, resulting in a small dynamic range of regulatory activity. Compared with binding at monomeric motifs, K50 HD's dimeric binding to P3 sequences is of much lower affinity (Wilson et al. 1993), and dimeric K50 HD motifs associated with terminal stage photoreceptor gene expression are made up of individually low-affinity half-sites. Thus, these dimeric K50 HD motifs are less active at low CRX concentration and remain responsive to higher and wider ranges of CRX concentrations. The requirement of CRX's cooperative dimerization for activity suggests that these low-affinity half-sites are poorly bound individually under the physiological CRX concentration and that cooperativity is crucial for stabilizing the binding complex. Many CRX-DAGs that are regulated by monomeric and dimeric K50 HD motifs encode proteins in the phototransduction pathway. Proteins in this pathway need to express robustly and remain dynamic in response to changes in ambient illumination. Thus, it is likely that combinational regulation of monomeric and dimeric K50 HD motifs imparts functional specification of CRX to regulate a subset of target genes that play specific physiological roles in photoreceptor biology. Collectively, precision in CRX–DNA interactions is important for not only the quantitative regulation but also the temporal control of photoreceptor gene expression.
Differential HD motif usage in photoreceptor development and diseases. (A) Q50 and K50 paired-class HDs prefer different monomeric and dimeric DNA motifs. (B,C) In postnatal photoreceptor precursors, regulatory elements with Q50 HD motifs are gradually closed (silenced) owing to a lack of interacting TFs. High-affinity monomeric K50 HD motifs respond to low concentration of CRX, drive chromatin remodeling in early-stage development, and plateau in their regulatory output at a later stage. Dimeric K50 HD motifs of individually low-affinity half-sites respond at high CRX concentration and are associated with chromatin remodeling and gene expression regulation at photoreceptor terminal differentiation. (D) In CrxK88N mutant retinas, CRXK88N’s ectopic binding impedes the silencing of regulatory elements with Q50 HD motifs. In CrxE80A mutant retinas, CRXE80A’s enhanced interactions with monomeric K50 HD motifs and defective cooperative binding at dimeric K50 HD motifs lead to accelerated chromatin remodeling in early-stage development but defective remodeling at terminal differentiation stage. (dLCA) Dominant Leber congenital amaurosis, (dCoRD) dominant cone-rod dystrophy.
Although the K50 HD motif division-of-labor model explains rod gene expression alternations in the CrxE80A retinas, it remains unclear why the subset of cone genes is significantly downregulated at both ages examined (Fig. 6B; Supplemental Fig. S6A). Because cones were born in CrxE80A mutant retinas, the loss of cell-type-specific gene expression in early postnatal development suggests defective differentiation (Zheng et al. 2023). Natively, cones and rods are generated from distinct pools of retinal progenitor cells that are inherently different in their competence (Hafler et al. 2012; Cepko 2015; Wang and Cepko 2016). It is conceivable that photoreceptor precursors generated from these distinct progenitor pools are different in their baseline epigenome architectures, which have a fundamental influence on the genetic programs required to reprogram the epigenome during cone and rod differentiation. In support of this prediction, 81.25% of the cone-enriched CRX-DAGs are associated with dimeric K50 HD motifs compared with only 55.45% of rod-enriched genes (Supplemental Fig. S5A), suggesting cone genes may be more dependent on CRX activity at dimeric K50 HD motifs. An alternative but not mutually exclusive model is that cone gene expressions are more sensitive to perturbations in CRX activity. In developing and mature mouse retinas, cones are dependent on a different repertoire of TFs than are rods for differentiation and functions (Swaroop et al. 2010; Forrest and Swaroop 2012; Emerson et al. 2013; Sapkota et al. 2014; Jean-Charles et al. 2018). Many rod-specific TFs collaborate with CRX in strongly activating rod gene expressions. Small perturbations in CRX activity may be dynamically compensated by CRX collaborating factors. In contrast, although many nuclear receptor family TFs have been identified to mediate M- versus S-cone subtype differentiation, these factors are dispensable for cone cell genesis, development, or survival in early postnatal ages (Forrest and Swaroop 2012). It is possible that CRX plays a major role in regulating general cone cell development and functions. The mouse retina is rod-dominant and thus is limited in the resolution of cone-related mechanistic understandings. Quantitative characterization of CRX molecular functions in a pure cone population and comparison with the CrxE80A model warrant further study to elucidate regulatory principles in early photoreceptor development and in CRX-linked dominant CoRD.
Besides the identification of two novel pathogenic mechanisms, our comparative analysis of the epigenome and transcriptome in WT and Crx mutant mouse retinas suggests two regulatory principles at important photoreceptor developmental transitions (Fig. 8). First, the transition from proliferating retinal progenitor cells to committed photoreceptors is accompanied by a shift from highly expressed Q50 to K50 paired-class HD TFs and from regulatory elements enriched with Q50 to those with high-affinity monomeric K50 HD motifs. Second, the transition from early- to late-stage photoreceptor development requires the utilization of dimeric K50 HD motifs in addition to monomeric K50 HD motifs. The first transition involves a sharp change in DNA-binding specificity, which likely confers sensitivity in newly postmitotic photoreceptor precursors to respond to low concentrations of CRX and quickly fix to a committed photoreceptor precursor status. This strategy may also ensure the rapid elimination of progenitor epigenetic features owing to a lack of interacting TFs. In contrast, the second principle implies a functional specialization of CRX at a specific set of target genes whose expressions need to remain dynamic and robust in mature photoreceptors. Related to the differential regulatory activities of different HD motifs, episomal reporter gene assays in the retinal explant system have shown that CRX-bound CREs containing multiple copies of high-affinity monomeric consensus K50 HD motifs are likely to act as silencers of gene expression (White et al. 2016; Friedman et al. 2021). Because these elements were tested outside of the native genomic context in an ex vivo system, whether they play a similar role in CRX's regulation of photoreceptor chromatin and gene expression in vivo warrants further study. Collectively, our findings support a unifying model in which differential CRX interactions with different HD motifs underlie cell-stage-specific chromatin remodeling and temporal gene regulation during photoreceptor development. A similar mechanism has been described for the SOX9 TF, in which its DNA-dependent cooperative dimerization is crucial for regulating genes for chondrogenesis but not for sex determination (Bernard et al. 2003). The examples of CRX and SOX9 suggest that some TFs have evolved distinct modes of DNA interactions that allow them to regulate diverse biological processes in different cellular contexts.
Our study here refines the CRX mechanistic model in photoreceptor development; expands our knowledge of the diverse mechanisms that CRX mutations lead to severe, dominant retinopathies; and lays the foundation for the future development of therapeutic strategies targeting different pathogenic mechanisms. Our CRX mechanistic model emphasizes the importance of considering interactions between coding CRX variants and noncoding variants in CRX binding sites in modifying clinical phenotypes. Our study also demonstrates, in addition to its unique biochemical properties (Wilson et al. 1993), that paired-class HD cooperative dimerization plays a crucial role in development and that its dysregulation can lead to distinct human diseases. The adaption of Coop-seq enables the unbiased identification of CRX HD cooperative dimerization as opposed to noncooperative cobinding, which is not easily separable in selection-based TF–DNA-binding assays. Our study provides hints toward understanding the structural basis of paired-class HD cooperative dimerization on palindrome DNA sequences (Wilson et al. 1995). Multiple missense mutations at the CRX E80 residue have been reported in dominant CoRD cases, including p.E80K (ClinVar VCV000099599), p.E80G (VCV000865803), and p.E80A (VCV000007416). Systematic investigation on how disease-associated variants affect CRX HD–DNA contacts and/or intramolecular contacts with other HD residues would guarantee new structural insights. Lastly, given that HD TF molecular mechanisms of action are conserved across evolution and in different tissues and organs, we envision our CRX study will also shed light on the study of other homeoproteins, which will hopefully lead to advances in medicine for the associated diseases.
Methods
Ethics statement
All procedures involving mice were approved by the animal studies committee of Washington University in St. Louis and performed under protocol 21-0414 (to S.C.). Experiments were carried out in strict accordance with recommendations in the guide for the care and use of laboratory animals of the National Institutes of Health, the Washington University policy on the use of animals in research, and the guidelines for the use of animals in visual research of the association for research in ophthalmology and visual sciences. Every effort was made to minimize the animals’ suffering, anxiety, and discomfort.
Coop-seq library preparation and EMSA
Escherichia coli expression and purification of GST-CRX HD peptides and preparation of Coop-seq libraries were performed as described previously (Chen et al. 2002; Zheng et al. 2023). The HD–DNA-binding reactions were performed in 1× CRX binding buffer (60 mM KCl, 25 mM HEPES, 5% glycerol, 1 mM DTT). The reaction mixtures were run in native 12% Tris-glycine PAGE gel (Invitrogen) at 160 V for 40 min at 4°C. The visible bands were excised from the gels. The extracted Coop-seq DNAs were extracted, purified, and PCR-amplified to tail on indexing barcodes and sequencing adapters. All Coop-seq samples were pooled and sequenced on a single 1 × 50 bp MiSeq run at the DNA Sequencing Innovation Laboratory at the Center for Genome Sciences & Systems Biology (CGS&SB; Washington University).
Determination of relative cooperativity with Coop-seq
For a combinatorial interaction of two (or more) protein species X and Y, and a particular DNA sequence, 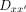 , which is a combination of two half-sites, x and
, which is a combination of two half-sites, x and 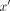 with a spacer, z, between them, the dimeric binding interaction can be diagrammed as
with a spacer, z, between them, the dimeric binding interaction can be diagrammed as (1)
where
(1)
where 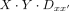 refers to the dimeric protein–DNA complex. Specifically, in our study, we are interested in single protein species P binding to a dimeric DNA sequence Dxx´. We can rewrite the equation above as
refers to the dimeric protein–DNA complex. Specifically, in our study, we are interested in single protein species P binding to a dimeric DNA sequence Dxx´. We can rewrite the equation above as (2)
(2)
In addition, in our system, the HD peptides (protein species P) are also competent to bind each half-site as a monomeric protein–DNA complex. There are two additional states of the DNA
sequence being bound: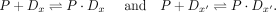 (3)
(3)
At equilibrium, the probability of a DNA molecule being in the unbound (U), monomerically bound (Bm), or dimerically bound (Bd) states (Fig. 2B) is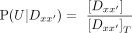 (4)
(4)
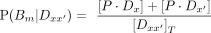 (5)
(5)
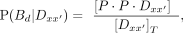 (6)
where […] refers to concentrations and
(6)
where […] refers to concentrations and 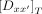 as the sum of all the states. The affinity of the protein P to a sequence Dx is defined as the association constant KA. The ratios of the probabilities above are related to the association constants as
as the sum of all the states. The affinity of the protein P to a sequence Dx is defined as the association constant KA. The ratios of the probabilities above are related to the association constants as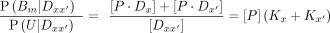 (7)
(7)
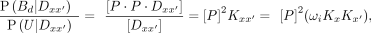 (8)
where ωi is the cooperativity and is unique to each dimeric sequence
(8)
where ωi is the cooperativity and is unique to each dimeric sequence 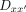 with different half-site combination and spacer. The cooperativity for each unique
with different half-site combination and spacer. The cooperativity for each unique 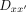 can be written as
can be written as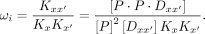 (9)
(9)
To obtain relative cooperativities relative to some reference sequence 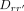 ,
,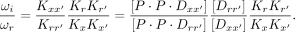 (10)
(10)
Because we are interested in comparing the cooperativity index ωi for half-site matched dimeric sequences with a 3 bp (P3) versus 5 bp (P5) spacer (Fig. 2C,D), the 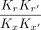 term can be neglected assuming the monomeric binding to half-sites is the same for the two sequence configurations. In a
binding reaction involving TF and a library of DNAs, the concentration of the bound and the unbound species is directly proportional
to the number of individual DNA molecules in each fraction obtained directly from the sequencing data. With sequencing counts
in each fraction, we can accurately estimate the ratios of concentrations from counts with the relationship
term can be neglected assuming the monomeric binding to half-sites is the same for the two sequence configurations. In a
binding reaction involving TF and a library of DNAs, the concentration of the bound and the unbound species is directly proportional
to the number of individual DNA molecules in each fraction obtained directly from the sequencing data. With sequencing counts
in each fraction, we can accurately estimate the ratios of concentrations from counts with the relationship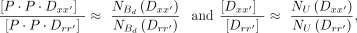 (11)
where NU denotes counts in the unbound fraction, and
(11)
where NU denotes counts in the unbound fraction, and 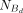 denotes counts in the dimerically bound fraction. We can then rewrite Equation 10 as
denotes counts in the dimerically bound fraction. We can then rewrite Equation 10 as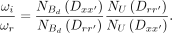 (12)
(12)
Coop-seq data analysis and cooperativity index calculation
The raw sequencing data for monomer and unbound (free) bands were included in our previous publication under NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) accession number GSE223658. The sequencing results were filtered and converted to count ratios following the Spec-seq analysis pipeline previously described (Zheng et al. 2023). Briefly, reads with any mismatch in the conserved regions were discarded, and sequences with fewer than 50 raw read counts were discarded. The relative cooperativity index ωi for each dimeric sequence was calculated following Equation 12. In Figure 2D, pair-wise relative cooperativity of half-site matched P3–P5 library members is presented. The relative cooperativity was ordered by hierarchical clustering using the Python package SciPy (v1.11.2) (Virtanen et al. 2020) with linkage method “complete” and distance metric “Euclidean.” In Supplemental Figure S2, A–D, a single reference sequence 5′-TAATGCGCTATTA-3′ was used to calculate relative cooperativity. We chose this sequence based on previous Spec-seq results that WT, E80A and K88N HDs all bind with reasonable affinity to either half-site. The full relative cooperativity index table for all library members can be found in Supplemental Table S2. The P3–P5 matched relative cooperativity index table sorted as in Figure 2D can be found in Supplemental Table S3.
ATAC-seq
For each genotype, three biological replicates and two retinas per replicate from one male and one female were pooled. The assay for transposase-accessible chromatin with sequencing was performed as previously published (Buenrostro et al. 2015). The quantity and quality of the ATAC-seq libraries were assayed using the Qubit 3 fluorometer (Invitrogen) and the Bioanalyzer (Agilent) prior to sequencing. All ATAC-seq sequencing libraries were pooled and sequenced on the Illumina NovaSeq 2000 platform (2 × 150 bp reads) with an average depth of 54 million reads at the Genome Technology Access Center at the McDonnell Genome Institute (GTAC@MGI; Washington University). The mm10 FASTA sequences for ATAC-seq peaks were obtained using R package BSgenome (v1.66.3) (https://bioconductor.org/packages/BSgenome). De novo motif enrichment analysis was performed with MEME-ChIP in the MEME suite (v5.5.2) (Bailey et al. 2015) using an order-one Markov background model and default parameters. Instances of K50 and Q50 HD monomeric and dimeric motifs were identified with FIMO in the MEME suite (v5.5.2) using an order-one Markov background model and ‐‐thresh 1.0 × 10−3.
MPRA library construction, electroporation, and sequencing
The library of 200mer oligonucleotides, each containing a 134 bp testing CRE sequence and a unique 10 bp barcode, was ordered directly from Twist Bioscience. The MPRA plasmid library was constructed using the 200mer oligonucleotides following published protocols (Hughes et al. 2018). For each retinal electroporation, 30 μg of MPRA plasmid library DNA per retina (three retinas per replicate, a total of three to four replicates per genotype) was used. The electroporated retinas were cultured for 8 days in the incubator (37°C, 5% CO2) before being harvested for RNA and DNA extraction using TRIzol reagent (Invitrogen). The MPRA sequencing libraries were prepared as previously described (Friedman et al. 2021). All MPRA sequencing libraries were pooled and sequenced on the Illumina NovaSeq2000 platform (2 × 150 bp reads) at the GTAC@MGI (Washington University). Libraries prepared from unelectroporated plasmid DNAs were sequenced to 50 million reads in two technical replicates (∼2500× depth). Libraries prepared from retinal explant extracted RNA (n = 3/4 per genotype) and DNA (n = 1 per genotype) were sequenced to an average depth of 30 million reads (∼1600× depth).
MPRA data analysis
The preprocessing of MPRA reads followed the pipeline previously described (Friedman et al. 2021). After dropping low-quality CREs, counts of individual RNA libraries were normalized by the average counts of the plasmid libraries to obtain a “raw activity score” for individual barcodes. Genotype average raw activity scores by unique testing CREs were calculated by averaging the barcode raw activity scores for each CRE. A coefficient of variation threshold of 1.0 was used to filter out CREs whose barcode activity varies greatly among genotype replicates. The “regulatory activity score” was calculated by normalizing the raw activity score of each CRE to the average raw activity score of all scrambled control CREs. In the MPRA library design, for each testing genomic CRE sequence (WT) that contains HD motif(s), one or more mutated CRE versions were generated by mutating the HD motif(s) (Supplemental Methods). The “HD motif activity score” was calculated as the “regulatory activity score” difference between a mutant CRE version and its matched genomic CRE sequence.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE256215. Customized scripts and any additional information required to reproduce the analysis in this paper are available as Supplemental Code and at GitHub (https://github.com/YiqiaoZHENG/CRXHD_epigenome.git [data visualization] and https://github.com/YiqiaoZHENG/CRXHD_mpra.git [dedicated MPRA design, sequencing library preparation, and data analysis]).
Competing interest statement
The authors declare no competing interests.
Acknowledgments
This work was supported by National Institutes of Health grants EY012543 to S.C., EY032136 to S.C., EY027784 to S.C. and B.A.C., and EY002687 to WU-DOVS; the Stein Innovation Award from Research to Prevent Blindness to S.C.; and unrestricted funds from Research to Prevent Blindness to WU-DOVS. We thank Mingyan Yang for technical assistance, Mike Casey from the Molecular Genetics Service Core for generating luciferase reporter assay plasmids and the MPRA oligo library, J. Hoisington-Lopez and M. Crosby from DNA Sequencing Innovation Laboratory at the Center for Genome Sciences and Systems Biology (CGS&SB), and the Genome Technology Access Center at the McDonnell Genome Institute (GTAC@MGI) for sequencing assistance. We also thank Mr. Artur Widlak for the generous gift from the Widłak Family CRX Research Fund.
Author contributions: S.C. and Y.Z. conceived the study. S.C. and G.D.S. supervised the study. S.C. and Y.Z. designed the experiments. Y.Z. performed all the experiments, data analysis, and visualization. G.D.S. assisted in Coop-seq data analysis. Y.Z. wrote the original draft. S.C., G.D.S., and Y.Z. revised the manuscript. All authors read and approved the final manuscript.
Footnotes
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.279340.124.
-
Freely available online through the Genome Research Open Access option.
- Received March 15, 2024.
- Accepted December 10, 2024.
This article, published in Genome Research, is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.