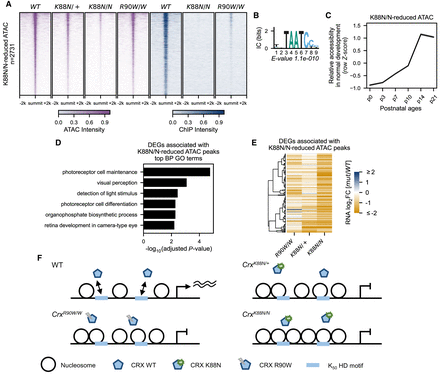
CrxK88N/+ retinas show defective chromatin remodeling at photoreceptor CREs enriched with K50 HD motifs. (A) Heatmaps depicting the normalized ATAC-seq or CRX ChIP-seq signal intensities at CrxK88N-reduced accessible ATAC-seq peaks. (B) PWM logo and enrichment significance E-value of the STREME de novo discovered HD motif. (C) Line plot showing the average developmental accessibility kinetics of CrxK88N-reduced ATAC-seq peaks. The developmental ATAC-seq data are from Aldiri et al. (2017). (D) Barchart showing biological process (BP) Gene Ontology (GO) term enrichment of differentially expressed genes adjacent to CrxK88N-reduced ATAC-seq peaks. (E) Heatmap comparing the P10 expression changes of CrxK88N-reduced ATAC-seq peaks associated genes in different Crx mutant retinas. The gene set is identical to that in D. (F) Schematics depicting chromatin remodeling defects at photoreceptor regulatory regions in the CrxK88N/+, CrxK88N/N, and CrxR90W/W retinas.











