Genome-wide phosphoacetylation of histone H3 at Drosophila enhancers and promoters
Abstract
Transcription regulation is mediated by enhancers that bind sequence-specific transcription factors, which in turn interact with the promoters of the genes they control. Here, we show that the JIL-1 kinase is present at both enhancers and promoters of ecdysone-induced Drosophila genes, where it phosphorylates the Ser10 and Ser28 residues of histone H3. JIL-1 is also required for CREB binding protein (CBP)-mediated acetylation of Lys27, a well-characterized mark of active enhancers. The presence of these proteins at enhancers and promoters of ecdysone-induced genes results in the establishment of the H3K9acS10ph and H3K27acS28ph marks at both regulatory sequences. These modifications are necessary for the recruitment of 14-3-3, a scaffolding protein capable of facilitating interactions between two simultaneously bound proteins. Chromatin conformation capture assays indicate that interaction between the enhancer and the promoter is dependent on the presence of JIL-1, 14-3-3, and CBP. Genome-wide analyses extend these conclusions to most Drosophila genes, showing that the presence of JIL-1, H3K9acS10ph, and H3K27acS28ph is a general feature of enhancers and promoters in this organism.
Cell differentiation in multicellular organisms requires the creation of complex spatio-temporal patterns of gene expression during development. The establishment and maintenance of these transcription blueprints entails the precise orchestration of a complex set of interactions among regulatory sequences and their binding proteins. Enhancers are one of these regulatory sequences capable of binding combinations of transcription factors that then recruit chromatin remodeling enzymes and/or interact with components of the Mediator complex or TFIID to help recruit RNA polymerase II (RNAPII) (Ong and Corces 2011). Transcription activation is accompanied by the establishment of a complex set of covalent histone modifications, such that the transcriptional status of a gene is determined by the presence of distinct modifications at enhancers and promoters. Well-documented are the interconnected phosphorylation, acetylation, and methylation of histones at enhancers and promoters that are necessary for proper transcription of genes (Berger 2007; Guenther et al. 2007; Heintzman et al. 2009).
Genome-wide studies have identified histone marks unique to enhancers, suggesting that the function of these regulatory elements may depend on combinations of specific covalent histone modifications. Enhancer regions are marked prior to activation by monomethylated H3K4 (H3K4me1) and acquire histone H3 acetylated on lysine 27 (H3K27ac) carried out by either CREBBP (CREB binding protein) or EP300 (also known as p300) (Heintzman et al. 2009). Studies of the FOSL1 gene have shown that phosphorylation of Ser10 of histone H3 preacetylated in Lys9 (H3K9acS10ph) by the PIM1 kinase at enhancer sequences promotes binding of 14-3-3 and facilitates acetylation of histone H4 on Lys16 (H4K16ac) by the histone acetyltransferase KAT8 (also known as MOF) (Zippo et al. 2009). Similarly, the MSK1/2 kinases phosphorylate histone H3S10 at the JUN (also known as c-Jun) and FOS (also known as c-Fos) genes in response to serum stimulation (Soloaga et al. 2003; Macdonald et al. 2005). Studies in Drosophila indicate that the JIL-1 kinase, which is the homolog of the mammalian MSK1/2 kinases, is also responsible for phosphorylation of H3S10 (Wang et al. 2001), but its precise role in transcription remains controversial (Cai et al. 2008). We have proposed a direct role for JIL-1 in transcription and shown that JIL-1 is recruited to the promoter of the hsp70 gene using standard chromatin immunoprecipitation (ChIP) assays and that H3S10ph is associated with other actively transcribed genes and is required for the release of RNAPII from promoter-proximal pausing (Nowak et al. 2003; Ivaldi et al. 2007). This mark then recruits 14-3-3 and the Elp3 histone acetyltransferase which acetylates H3K9 and is necessary for the release of the paused polymerase (Karam et al. 2010).
In this study, we analyze the role of JIL-1 in some of the events that take place before and after initiation of transcription and are necessary to proceed to productive elongation upon gene activation. To overcome concerns affecting the use of immunofluorescence techniques using acid-fixed polytene chromosomes (Cai et al. 2008), we instead use standard Chip and ChIP-seq assays using antibodies previously validated by ENCODE and modENCODE. We find that enhancers and promoters of ecdysone-induced genes undergo phosphorylation in the Ser10 and Ser28 residues of histone H3 by the JIL-1 kinase. The same histone tails are also acetylated at Lys9 and Lys27. These modifications are able to recruit 14-3-3, which we demonstrate is necessary to achieve a stable interaction between enhancer and promoter. We then extend these findings by examining the localization of these proteins genome-wide and conclude that all or most Drosophila enhancers and promoters contain JIL-1, H3K9acS10ph, and H3K27acS28ph. These results may offer important insights into the mechanisms used by cells to establish and/or maintain interactions between distant regulatory sequences in the genome.
Results
JIL-1 is responsible for H3S28 phosphorylation during transcription in D. melanogaster
To investigate whether H3S28 phosphorylation is involved in transcription in Drosophila cells and JIL-1 might be the kinase responsible for carrying out this modification, we examined polytene chromosomes from third instar larvae by immunofluorescence microscopy using antibodies specific to H3S28ph. The results of these experiments show that H3S28ph is present in interbands of polytene chromosomes, which contain actively transcribed genes, and appears at the heat shock puffs upon temperature elevation (Supplemental Fig. S1). Polytene chromosomes from JIL-1 mutant larvae lack H3S28ph at the heat shock genes after exposure to elevated temperature. Lysates from salivary glands of JIL-1 mutants also show a dramatic loss of H3S28ph (Supplemental Fig. S1C). Furthermore, in vitro kinase assays using JIL-1 immunoprecipitated from cell extracts show abundant phosphorylation of H3S28 (Supplemental Fig. S1D). These data suggest that JIL-1 is the kinase responsible for H3S28 phosphorylation upon transcriptional activation in Drosophila.
JIL-1 phosphorylation of H3S10 and H3S28 at promoters and enhancers is necessary for transcriptional activation of ecdysone-responsive genes
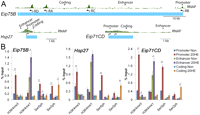
To better understand the role of JIL-1 in transcription activation, we examined the presence of this protein at genes induced by the steroid hormone 20-hydroxyecdysone (20-HE) using ChIP experiments in Kc167 cells treated with this hormone. We analyzed three different ecdysone-induced genes, Eip75B, Hsp27, and Eip71CD, whose structure and organization are shown in Figure 1A (Riddihough and Pelham 1987; Cherbas et al. 1991; Bernardo et al. 2009). Interestingly, all three genes contain a significant amount of polymerase at the 5′ end of the gene despite relatively low levels inside the gene, suggesting that these genes have a paused polymerase (Fig. 1A). Chromatin immunoprecipitation assays performed on both untreated cells and cells treated with 20-HE for 3 h demonstrate that the predicted enhancers for all three genes contain high levels of H3K4me1, a characteristic mark of enhancers, which increases upon activation. There is a small amount of initiated polymerase (RNAPII S5ph) at all three genes, in agreement with the fact that they are transcribed at low levels in untreated Kc cells (Gauhar et al. 2009; Wood et al. 2011). After induction, H3K4me3 increases at the promoter, while an increase of elongating RNA polymerase (RNAPII S2ph) is detected at promoters and coding regions but not at enhancers (Fig. 1B). These results verify that these promoter/enhancer pairs have the expected histone modifications after transcription activation.
Phosphorylation of H3S10 and H3S28 by JIL-1 at promoters and enhancers is necessary for transcriptional activation of ecdysone responsive genes. (A) Structure of three early 20-HE responsive genes with previously characterized enhancers showing the location of promoter, enhancer, and coding sequences used in ChIP analyses. ChIP-chip (modENCODE) profile of RNAPII large subunit indicated in green above each gene. (B) ChIP analysis of Kc167 cells treated for 3 h with ethanol (Non) or 20-HE (20HE). Antibodies used for pull-down are indicated along the x-axis. (Ser5ph) RNAPII phosphorylated on serine 5 of CTD, (Ser2ph) RNAPII phosphorylated on serine 2 of CTD. DNA was quantified by real-time PCR using primers designed to amplify the promoter, enhancer, and coding regions of each of the three genes.
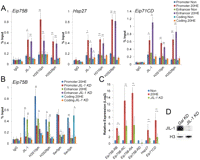
We then examined whether JIL-1 is recruited to enhancers, promoters, and coding regions upon gene activation after 20-HE treatment. We also analyzed the presence of H3S10ph and H3S28ph, the two modifications carried out by JIL-1, at these regions when transcription is turned on. ChIP experiments using antibodies to JIL-1, H3S10ph, and H3S28ph were performed with and without 20-HE treatment of Kc167 cells. Results indicate that JIL-1 is recruited to both enhancers and promoters after 20-HE treatment and that H3S10ph and H3S28ph increase at these two sequences concomitant with the presence of JIL-1 (Fig. 2A). In order to confirm that JIL-1 is responsible for the phosphorylation of H3 detected at the enhancers and promoters of the ecdysone responsive genes, we knocked down this enzyme in Kc167 cells using dsRNA and performed the same ChIP experiments after treatment with 20-HE for 3 h (Fig. 2B). The JIL-1 knockdown cells show a severe growth delay and, at higher levels of knockdown, a lethal phenotype. The ChIP data demonstrate that loss of JIL-1 results in loss of H3S10ph and H3S28ph from both the promoter and enhancer upon induction when compared to the Gal knockdown used as a control. Additionally, the levels of initiated and elongating polymerase at the promoter and coding regions suggest that JIL-1 knockdown interferes with transcription elongation regardless of normal initiation of polymerase at the promoter (Fig. 2B). A complete knockdown of JIL-1, manifested by cell growth defects, is necessary to observe these effects on transcription elongation. These data confirm earlier results suggesting that JIL-1 functions after transcription initiation but before elongation (Ivaldi et al. 2007). To confirm the transcription defects caused by lack of JIL-1, we isolated mRNA, followed by reverse transcription to measure transcript levels of the Eip75B gene in JIL-1 knockdown versus Gal knockdown cells. Eip75B encodes four transcripts named Eip75B–RA, –RB, –RC, and –RD (Fig. 1A); the first three transcripts undergo a large fold-increase in expression upon hormone treatment, whereas the Eip75B–RD transcript shows little or no increase. In cells lacking JIL-1, all three inducible transcripts show significantly lower expression levels (Fig. 2C). Therefore, JIL-1 is responsible for the H3S10ph and H3S28ph modifications observed at both the enhancer and promoter, and lack of these modifications in JIL-1 knockdown cells correlates with failure to activate transcription.
JIL-1 is required for phosphorylation at enhancers and promoters upon ecdysone transcriptional activation. (A) ChIP analysis of Kc167 cells treated for 3 h with ethanol (Non) or 20-HE (20HE). Antibodies used for pull-down are indicated along the x-axis. (Ser5ph) RNAPII phosphorylated on serine 5 of CTD, (Ser2ph) RNAPII phosphorylated on serine 2 of CTD. DNA was quantified by real-time PCR using primers designed to amplify the promoter, enhancer, and coding regions of each of the three genes. (B) ChIP analysis in Kc167 cells in which JIL-1 or Gal expression was inhibited by RNAi. Cells were treated with 20-HE for 3 h. Antibodies used are indicated along the x-axis. (Ser5ph) RNAPII phosphorylated on serine 5 of CTD, (Ser2ph) RNAPII phosphorylated on serine 2 of CTD. DNA was quantified by real-time PCR, and results are reported as percentage of input. (C) RNA expression analysis in cells treated with dsRNAs corresponding to the JIL-1 or Gal genes. Cells were treated with ethanol (Non) or 20-HE (20HE) for 3 h. RNA levels were determined by qPCR using primers specific to each of the four Eip75B transcripts. All samples were normalized to the mitochondrial gene myt:Col and the Non sample was set to 1 for comparison. (D) Western analysis of dsRNA-treated cells showing undetectable levels of JIL-1. Error bars represent the standard deviation of the mean of 3 biological replicates. (*) P < 0.05, (**) P < 0.01.
CBP acetylation at promoters and enhancers is necessary for transcription activation of ecdysone-responsive genes
H3K27ac is a characteristic mark of active enhancers and is deposited by either CBP or EP300 in mammals (Visel et al. 2009). In Drosophila, there is no ortholog of EP300, and CBP is the only reported acetyltransferase responsible for catalyzing this acetylation event (Tie et al. 2009). H3S28 phosphorylation facilitates loss of the repressive methylation mark H3K27me3 in the neighboring residue and subsequent acetylation of the same lysine (Tie et al. 2009; Lau and Cheung 2011). Acetylation of H3K9 is enhanced by 14-3-3, a phospho-serine binding protein, via recruitment of the ELP3 acetyltransferase (Karam et al. 2010). D. melanogaster has two isoforms of 14-3-3, 14-3-3ζ and 14-3-3ɛ, both of which have been shown to bind H3S10ph and H3S28ph (Macdonald et al. 2005). Coimmunoprecipitation experiments with either H3S10ph or H3S28ph antisera demonstrate a strong interaction with 14-3-3 (Supplemental Fig. S2). We hypothesize that a similar process may take place at enhancers, with 14-3-3 binding H3S10ph/H3S28ph to recruit CBP and acetylate H3K27.
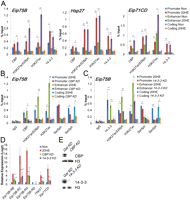
To confirm binding of CBP and 14-3-3 at enhancers and promoters and acetylation of H3K27 upon transcriptional activation, we performed ChIP assays at the three selected ecdysone-responsive genes. The 14-3-3 antibody detects both isoforms of Drosophila 14-3-3. Results show that CBP and 14-3-3 are recruited to both enhancers and promoters after 20-HE treatment. In addition, H3K27ac undergoes a significant increase at these two regulatory sequences upon gene activation (Fig. 3A). ChIP analyses using antibodies against the dual modification H3K27acS28ph suggest that acetylation and phosphorylation occur on the same histone tail. These results indicate that most of the increase in H3K27ac detected upon transcription activation takes place in the tail of H3 carrying the H3S28ph mark at these enhancers and promoters.
CBP acetylation and 14-3-3 recruitment at promoters and enhancers is necessary for transcription activation of ecdysone-responsive genes. (A) ChIP analysis of Kc167 cells treated for 3 h with ethanol (Non) or 20-HE (20HE) was performed using the antibodies indicated along the x-axis. (Ser5ph) RNAPII phosphorylated on serine 5 of CTD, (Ser2ph) RNAPII phosphorylated on serine 2 of CTD. DNA was quantified by real-time PCR using primers designed to amplify the promoter, enhancer, and coding regions of each of the three ecdysone-induced genes. (B) ChIP analysis of Kc167 cells nontreated and treated with CBP dsRNA compared to Gal dsRNA. The cells were also incubated with 20-HE for 3 h, and chromatin was immunoprecipitated using the antibodies indicated along the x-axis. DNA was quantified by real-time PCR, and the result is reported as a percent of the input at the Eip75B gene. (C) Similar to panel B, but cells were treated with dsRNA to 14-3-3. (D) RNA expression analysis in cells treated with dsRNAs corresponding to the CBP, 14-3-3, or Gal genes. Cells were treated with ethanol (Non) or 20-HE (20HE) for 3 h. RNA levels were determined by qPCR using primers specific to each of the four Eip75B transcripts. All samples were normalized to the mitochondrial gene myt:Col and the Non sample was set to 1 for comparison. (E) Western analysis of lysates from cells treated with CBP (top) or 14-3-3 (bottom) dsRNAs. Levels of either protein are undetectable. Error bars represent the standard deviation of the mean of 3 biological replicates. (*) P < 0.05, (**) P < 0.01.
To confirm that CBP is the acetyltransferase responsible for acetylation of H3K27 during the ecdysone response, ChIP was performed in CBP knockdown cells (Fig. 3E) at the Eip75B gene. Down-regulation of CBP results in a dramatic decrease of H3K27ac and H3K27acS28ph at both promoter and enhancer (Fig. 3B). Lack of CBP had no effect on the amount of initiating RNAPII but caused a reduction in the levels of elongating polymerase. The same experiments were then performed using knockdown cells for both isoforms of 14-3-3 (Fig. 3E). Lack of 14-3-3 results in a reduction of H3K27ac upon hormone induction. However, there is not complete loss of H3K27ac, suggesting that the role of 14-3-3 in CBP activity may be partially redundant (Fig. 3C). Normal levels of initiated polymerase are present at the promoter of Eip75B in both Kc cells treated with CBP or 14-3-3 dsRNAs, but loss of the elongating polymerase suggests that these proteins may function at the promoter-proximal pause step. The loss of elongating polymerase is confirmed by the significant decrease of all three 20-HE-induced Eip75B transcripts in cells lacking CBP or 14-3-3 (Fig. 3D). Neither mutation of CBP or 14-3-3 has an effect on the levels of H3S28ph (Supplemental Fig. S3). From these data, we can conclude that CBP activity, enhanced by 14-3-3, can be detected at both enhancers and promoters and is necessary for transcription elongation.
14-3-3 recruitment and H3K27 acetylation are dependent on JIL-1 kinase at enhancers and promoters before release of polymerase from promoter-proximal pause
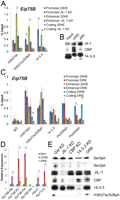
If the hypothesis that H3K27 acetylation is facilitated by H3S28 phosphorylation and subsequent 14-3-3 recruitment is correct, then loss of H3S28ph should result in a decrease of H3K27ac. To test this, we knocked down JIL-1 with dsRNAs and examined the presence of H3K27ac and 14-3-3 using ChIP. We can conclude from the dramatic decrease of both 14-3-3 and H3K27ac that the activity of CBP and 14-3-3 recruitment is dependent on JIL-1 at enhancers of ecdysone-responsive genes (Fig. 4A); although there is also a significant decrease in H3K27acS28ph and 14-3-3 in promoter regions, the difference in H3K27ac before and after ecdysone induction is not statistically significant. Coimmunoprecipitation experiments demonstrate that CBP and JIL-1 physically interact and the interaction is facilitated by phosphorylation (Fig. 4B). JIl-1 interacts with 14-3-3ɛ (top band); this is probably not the same 14-3-3 molecule associated with histones, since histones interact with both the ɛ and ζ isoforms (Supplemental Fig. S2).
14-3-3 recruitment and H3K27 acetylation are dependent on JIL-1 kinase at enhancers and promoters. (A) Kc167 cells treated with dsRNAs to Gal or JIL-1 were incubated with 20-HE for 3 h and subjected to ChIP analysis using the antibodies indicated along the x-axis. DNA was quantified by real-time PCR and reported as a percent of the input. (B) Kc167 cell lysates with and without phosphatase inhibitors (PPI) were immunoprecipitated using antibodies against JIL-1, loaded 1:10 with respect to input, and subjected to Western analysis using antibodies against JIL-1, CBP, and 14-3-3. (C) Using Kc167 cells pretreated for 5 min with 100 μM DRB followed by 3-h treatment with 20-HE (20HE), ChIP was performed using the antibodies indicated along the x-axis and quantitated by real-time PCR using primers designed to amplify the promoter, enhancer, and coding regions of the Eip75B gene. (D) Relative expression analysis of the DRB-treated cells compared with normal 20-HE induction using primers specific to each of the four transcripts of Eip75B. All samples were normalized to mitochondrial gene product myt:Col and the Non sample set to 1 for comparison. Error bars represent the standard deviation of the mean of 3 biological replicates. (E) Western analysis of lysates from Kc167 cells treated with dsRNAs corresponding to the JIL-1, CBP, 14-3-3, or Gal genes; cells were pretreated with DRB for 5 min and with 20-HE for 3 h. Error bars represent the standard deviation of the mean of 3 biological replicates. (*) P < 0.05, (**) P < 0.01.
Western blots were performed on lysates of all the knockdown cells to ensure that there is not a loss of any of the other proteins as a secondary effect of inhibition of transcription in these cells. Results show that levels of JIL-1 are normal in the 14-3-3 knockdown (Fig. 4E). To further test a possible effect of inhibition of transcription on protein levels, cells were treated with DRB, an inhibitor of the positive transcription elongation factor b (P-TEFb). DRB-treated Kc167 cells show normal levels of H3K27acS28ph as measured by Western blots (Fig. 4E) or ChIP analysis of the Eip75B gene upon 20-HE induction (Fig. 4C) but fail to activate transcription of any of the Eip75B gene isoforms when measured by quantitative PCR (Fig. 4D). These results demonstrate that H3S28 phosphorylation and H3K27 acetylation take place before the release of the polymerase, the step inhibited by DRB treatment.
JIL-1, 14-3-3, and CBP are required for enhancer-promoter interactions
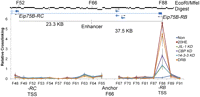
We then used chromosome conformation capture (3C) (Dekker et al. 2002) to test the possibility that physical contacts between an enhancer and promoter of the Eip75B gene depend on the presence of the JIL-1, CBP, or 14-3-3 proteins. The specific enhancer is located 37.5 kb downstream from the Eip75B-RB promoter and has been previously shown to be responsible for activating expression of this gene (Bernardo et al. 2009). The results show no interaction between the enhancer and the Eip75B-RB promoter in Kc167 cells (Fig. 5). After a 3 h ecdysone treatment, when expression of the Eip75B-RB gene is induced, we observe a dramatic increase in the interaction between the enhancer and the promoter of the gene (Fig. 5). To determine the role of H3K27 acetylation and H3S10 and H3S28 phosphorylation in the establishment of these interactions, we next performed 3C on CBP or JIL-1 knockdown cells. In both cases, lack of the CBP or JIL-1 proteins results in a decrease in the strength or frequency of the interactions between the enhancer and the promoter, suggesting that the presence of the proteins themselves or the histone modifications they perform correlate with the establishment and/or maintenance of contacts between these two sequences. Since the role of H3S10ph and H3S28ph is to recruit 14-3-3 and this protein normally plays a role as a scaffold of large protein complexes, we examined whether enhancer-promoter interactions are affected in cells lacking both isoforms of 14-3-3. The results show a dramatic decrease in enhancer-promoter contacts detected by 3C in cells treated with ecdysone and lacking 14-3-3, suggesting an essential role for this protein in bringing together these two sequences in order to activate transcription (Fig. 5).
JIL-1, 14-3-3, and CBP are required for enhancer-promoter interactions. 3C analysis of Kc167 cells after Gal knockdown and treatment with ethanol (Non) or 20-HE (20HE) for 3 h was performed in normal cells as well as cells in which the expression of the JIL-1, CBP, 14-3-3, or Gal genes was inhibited by treatment with the corresponding dsRNAs. 3C analysis was also done on cells treated with DRB for 5 min. Cross-linking efficiencies are reported after normalizing to a BAC clone with an insert of 170 kb containing the entire Eip75B locus. A restriction digest of this DNA gives 112 fragments numbered 1 to 112, beginning with the first fragment of the BAC. Error bars represent the standard deviation of the mean of 3 biological replicates.
Since knockdown of JIL-1, CBP, or 14-3-3 inhibits transcription, it is possible that the observed effects on enhancer-promoter interactions are indirect and caused by lack of expression of other factors important for transcription. To discard this possibility, we examined the effect of DRB on promoter-enhancer interactions at the Eip75B-RB gene using 3C. DRB-treated cells, however, are able to achieve contacts between the promoter and the enhancer that are similar in frequency to those in untreated cells, even though transcriptional elongation of the gene is lost (Fig. 5). These data suggest that promoter-enhancer contacts in the Eip75B gene depend on histone modifying enzymes that recruit 14-3-3, which, in turn, may serve as a physical bridge that stabilizes these interactions.
JIL-1 and phosphoacetylated H3 are present at promoters genome-wide, and their quantity correlates with transcription levels
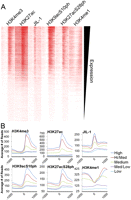
In order to investigate whether the presence of JIL-1, H3K9acS10ph, and H3K27acS28ph at promoters is specific for ecdysone-inducible genes or is a general phenomenon, we carried out ChIP-seq experiments. Genome-wide data sets were generated in Kc cells not treated with ecdysone for H3K4me3, H3K4me1, H3K27ac, JIL-1, H3K9acS10ph, and H3K27acS28ph. Heat maps of each modification around the TSS sorted by transcript levels show a correlation between the presence and amounts of JIL-1, H3K9acS10ph, and H3K27acS28ph at gene promoters and levels of transcription (Fig. 6A). A peak of JIL-1 is directly present over the TSS and drops within the first 200 bp inside the gene (Fig. 6B). This distribution is consistent with the proposed association of JIL-1 with paused RNAPII. Examples of the distribution of JIL-1 with respect to genes based on these ChIP-seq data and in comparison with ChIP-chip data sets obtained by modENCODE are shown in Supplemental Figure S4. Both H3K9acS10ph and H3K27acS28ph have very similar profiles as H3K4me3, suggesting these histone modifications occur on the same nucleosomes, the peak of which is located at the first nucleosome downstream from the TSS (Fig. 6A). These results suggest that JIL-1 is present at the promoter region of actively transcribed genes and correlates with transcript levels, supporting the idea that JIL-1 plays a direct role in the transcription process.
JIL-1 and histone phosphoacetylation at promoters correlate with transcript levels genome-wide. (A) RNA expression levels for Kc167 cells were obtained from modENCODE, and the TSSs were sorted by expression level from highest to lowest and given up/downstream orientation according to strand. The total number of reads is plotted for each ChIP-seq data set in 20-bp bins covering 1 kb upstream of and downstream from each TSS and viewed in Java Treeview. (B) All TSSs were broken down into five equal bins according to expression levels, and the average number of reads for each 20-bp region for all the TSSs in that bin is plotted 1 kb upstream and downstream for each bin.
JIL-1 and phosphoacetylated H3 are found at enhancers genome-wide prior to activation of transcription
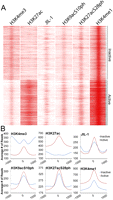
To investigate a possible role for JIL-1 at enhancers genome-wide, we first identified regions with significant enrichment for H3K4me1 (15,369) and then subtracted any that were within 1 kb of any TSS (10,655). Heat maps generated for all the data sets were clustered to identify patterns between sets. Clustering appeared to be driven by the presence or absence of H3K27ac (Fig. 7A). H3K27ac has been identified as a histone modification associated with active enhancers, and it is the best-characterized measure of enhancer activity (Heintzman et al. 2009; Creyghton et al. 2010; May et al. 2012). JIL-1, H3K9acS10ph, and H3K27acS28ph appear to be present at gene enhancers irrespective of the levels of H3K27ac. Nevertheless, the cluster of genes containing high levels of H3K27ac appears to also have higher levels of JIL-1 and both phosphoacetylated forms of H3 (Fig. 7B). Overall, the results suggest that JIL-1 and phosphoacetylated histone H3 are present at enhancers prior to activation and that their levels increase when enhancers are active.
JIL-1, H3K9acS10ph, and H3K27acS28ph are present at enhancers. Enhancers were selected by the presence of H3K4m1 and clustered into two clusters using Cluster 3.0. (A) The total number of reads is plotted for each ChIP-seq data set in 20-bp bins covering 1 kb upstream and downstream from the H3K4me1 peak summit and viewed in Java Treeview. (B) The average number of reads plotted for every H3K4me1 peak in the inactive or active cluster in 20-bp bins covering 1 kb upstream and downstream from the summit of H3K4me1.
Discussion
Activation of transcription in higher eukaryotes requires the interaction between transcription factors bound to distal enhancers and proteins present at the promoter. Recent findings indicate that enhancers contain a variety of histone modifications that change during the establishment of specific cell lineages suggesting that these sequences may play a more complex role in transcription than previously thought (Ong and Corces 2011). Given the presence of common as well as specific histone marks at enhancers and promoters, it is tempting to speculate that epigenetic modifications at these sequences serve to integrate various cellular signals required to converge in order to activate gene expression. Results described here support this hypothesis, demonstrating that the proteins that carry out these histone modifications are necessary to establish enhancer-promoter contacts and activate transcription of ecdysone-inducible genes.
The execution of this process in Drosophila requires the recruitment of JIL-1 by mechanisms that are not well understood. Although the direct involvement of JIL-1 in the transcription process has been brought into question due to the failure to observe recruitment of JIL-1 to heat shock genes in polytene chromosomes (Cai et al. 2008), results presented here clearly indicate that JIL-1 affects transcription at different steps in the transcription cycle. At the promoter region, phosphorylation of H3S10 by JIL-1 results in the recruitment of 14-3-3 and, subsequently, histone acetyltransferases Elp3 and MOF (Karam et al. 2010). Here, we find that JIL-1 is also able to phosphorylate H3S28 at both promoters the enhancers. The establishment of the H3K9acS10ph and H3K27acS28ph modifications correlates with the recruitment of 14-3-3 to enhancers and promoters of ecdysone-induced genes. 14-3-3 has been implicated in numerous cellular processes, where it functions as a scaffold protein (Morrison 2009). 14-3-3 is found as dimers and multimers; each monomer is capable of binding two targets and can mediate and stabilize interactions between two phosphoproteins. Additionally, acetylation facilitates the dimerization of 14-3-3 molecules and their ability to bind certain substrates (Tzivion et al. 1998; Choudhary et al. 2009). Binding assays have demonstrated that 14-3-3 interacts weakly with H3 tail peptides phosphorylated at S10 and S28, but strong binding is detected if the peptide is both phosphorylated and acetylated on the neighboring lysine residues (Walter et al. 2008; Winter et al. 2008). Given the ability of 14-3-3 to serve as a scaffold for large protein complexes, its demonstrated interactions with H3K9acS10ph and H3K27acS28ph and the presence of these two modifications at enhancers and promoters, it is possible that contacts between these two sequences are stabilized by 14-3-3. This hypothesis is supported by 3C experiments indicating that induction of transcription of the Eip75B gene is accompanied by strong enhancer-promoter interactions. These interactions are lost in JIL-1, CBP, and 14-3-3 knockdown cells. Since these proteins act several steps downstream from transcription factor binding in the pathway leading to enhancer-promoter contacts, and loss of these proteins results in the abolishment of these contacts, it appears that these proteins, rather than specific transcription factors, may be responsible for enhancer promoter interactions at the ecdysone-inducible genes.
Genome-wide studies using ChIP-seq clearly show the presence of JIL-1, H3K9acS10ph, and H3K27acS28ph at enhancers and promoters of most Drosophila genes. There is a clear correlation between the amount of JIL-1, H3K9acS10ph, and H3K27acS28ph at promoters and the level of transcripts associated with the gene. These three marks are also present at enhancers defined by the occurrence of H3K4me1 and H3K27ac, suggesting that the JIL-1 kinase is a regulator of histone dynamics at enhancers and promoters genome-wide. JIL-1, H3K9acS10ph, and H3K27acS28ph are found at low levels at enhancers before activation, which then increase in intensity and drop in baseline when found in combination with H3K27ac, a mark of active enhancers. These conclusions are different from those previously published examining the role of JIL-1 in transcription and dosage compensation (Regnard et al. 2011). The authors from this study conclude that JIL-1 binds active genes along their entire length and that the levels of JIL-1 are not associated with levels of transcription. The differences in the conclusions may be due to the different cell lines used—male S2 cells versus female Kc cells—and the emphasis of the analysis by Regnard et al. on the expression of dosage-compensated genes in the male X-chromosome, which may contain JIL-1 throughout the genes as a consequence of their regulation at the elongation step. In addition, the study by Regnard et al. used ChIP-chip on custom tiling arrays of the X chromosome plus cDNA arrays containing the whole genome. This strategy may bias the conclusions and suggest the presence of JIL-1 in the coding region of genes rather than at enhancers and promoters.
Results presented here extend the previous list of histone modifications characteristic of active enhancers to include H3K9acS10ph and H3K27acS28ph. Enhancers tend to be cell type-specific and are determined during differentiation with the characteristic H3K4me1 modification (Taberlay et al. 2011). It is unclear how these regions are designated before activation and what keeps them in a poised state ready for activation upon receiving the proper signal from the cell. It is tempting to speculate that the presence of JIL-1 at enhancers prior to activation might play a role in maintaining the enhancer in this poised state. An important question for future studies is the mechanistic significance of the looping between enhancers and promoters in order to achieve transcription activation. One interesting possibility is that various signaling pathways in the cell contribute to building epigenetic signatures at enhancers and promoters in the form of histone acetylation and/or phosphorylation of various Lys/Ser/Thr residues. Acetylation marks at enhancers and promoters may then cooperate to recruit BRD4 (FS(1)H in Drosophila), which contains two bromodomains each able to recognize two different acetylated Lys residues (Vollmuth et al. 2009). The requirement for acetylation of histones at enhancers and promoters in order to recruit Brd4 would ensure that several different signaling events have taken place before recruitment of P-TEFb by BRD4 can release RNAPII into productive elongation.
Methods
Treatment of cells with 20-HE, dsRNA, and DRB
Kc167 cells were grown in CCM3 serum-free insect media (HyClone SH30065.01) at 25°C. For RNAi treatment, cells were plated at 0.5 × 106 cells/mL and incubated with dsRNA targeting either the JIL-1, CBP, or 14-3-3ζ and 14-3-3ɛ genes (Supplemental Table S1) using Cellfectin II reagent (Invitrogen 10362-100). After addition of dsRNA, cells were grown for 4 d, replated at 2 × 106 cells/mL, retreated with dsRNA as before and grown for four additional days. For control RNAi treatments, the same procedure was performed using dsRNA targeting the Gal gene, but incubations were 2 d after each treatment instead of 4 d to collect a similar number of cells. All experiments labeled “Non” in the figures were treated with ethanol (the solvent used to prepare 20-HE solutions) for 3 h, and all experiments labeled 20-HE, JIL-1 KD, CBP KD, and 14-3-3 KD were treated with 20-HE (Sigma H5142) at a final concentration of 5 × 10−7 M for 3 h. For DRB (5,6-dichlorobenzimidazole 1-β-D-ribofuranoside; Sigma D1916) treatment, cells grown to 2 × 106 cells/mL were pretreated for 5 min at a concentration of 100 μM; 20-HE was then added to the media for an additional 3-h incubation as described above.
Antibodies and co-IP experiments
All antibodies to histone modifications used in the studies described here have been tested by modENCODE and shown to specifically recognize appropriate epitopes (http://compbio.med.harvard.edu/antibodies/). The following antibodies were used in this study. Histone H3 (Abcam ab1791), H3K4me3 (Abcam ab8580), H3K4me1 (Abcam ab8895), RNAPII S5ph (Covance MMS-134R), RNAPII S2ph (Covance MMS-129R), H3S28ph (Millipore 07-145), H3K27acS28ph (Millipore 05-896), H3K27ac (Abcam ab4729), JIL-1 (Karam et al. 2010), H3S10ph (Millipore 05-806), 14-3-3 (Santa Cruz Biotechnology sc-629), CBP (M. Mannervik, Stockholm University). Specificity of the antibodies against H3K27ac and H3K27acS28 was examined using dot blots of the appropriate peptides, and it is shown in Supplemental Figure S5. For immunoprecipitation experiments, 2 × 106 Kc167 cells were lysed in cell lysis buffer (5 mM PIPES [pH 8.0], 85 mM KCl, 0.5% TX-100), nuclei were spun down for lysis in 1 mL RIPA buffer containing protease inhibitor complex (Roche 04 693 159 001), and briefly sonicated to ensure lysis. Nuclear lysates were treated with 5 μl DNase I (Sigma D5319) plus 6 mM MgCl2 at 25°C for 20 min to obtain single nucleosomes. Antibodies were incubated in lysate at 4°C for 6 h and precipitated with a 50/50 mixture of Protein A and Protein G beads. After four washes in RIPA buffer, bound proteins were eluted in 1× Laemmli buffer and analyzed by Western blot.
ChIP and 3C analysis
ChIP experiments were performed on 2 × 107 Kc167 cells as described previously (Wood et al. 2011).
ChIPs were quantified as percent of input on a standard curve using SYBR real-time PCR. n = 5 biological replicates of the Gal knockdown samples labeled Non and 20HE, n = 3 biological replicates for JIL-1 KO, CBP KO, 14-3-3 KO, and DRB samples. For ChIP-seq experiments, libraries were sequenced on an Illumina GAII or HiSeq system, and reads were aligned to the Drosophila dm3 genome using Bowtie v0.12.7. Peak files and wiggle files were generated using MACS v1.4.1. 3C-qPCR analysis was done as described (Wood et al. 2011). Primers used for anchor and bait fragments can be found in Supplemental Table S2. Error bars represent the standard deviation of the mean of 3 biological replicates.
Expression analysis
Cells (1 × 106) were collected, RNA extraction was carried out using the RNeasy kit (Qiagen), and the RNA was reverse transcribed to cDNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Transcript specific primers spanning exon junctions for the Eip75B gene are as described (Wood et al. 2011). All samples were normalized to mitochondrial gene product myt:Col and the Non sample set to 1 for comparison. Error bars represent the standard deviation of the mean of 3 biological replicates for the JIL-1, CBP, and 14-3-3 knockdowns and 5 biological replicates of the Gal knockdown.
Data access
ChIP-seq data are deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE36374.
Acknowledgments
We thank Mattias Mannervik for the gift of CBP antibody and members of the Corces Lab for invaluable discussions. We also thank The Genomic Services Lab at the HudsonAlpha Institute for Biotechnology for their help in performing Illumina sequencing of ChIP-seq samples. This work was supported by U.S. Public Health Service Award GM35463 from the National Institutes of Health.
Footnotes
-
↵1 Corresponding author.
E-mail vcorces{at}emory.edu.
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.136929.111.
- Received December 22, 2011.
- Accepted April 4, 2012.
This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date (see http://genome.cshlp.org/site/misc/terms.xhtml). After six months, it is available under a Creative Commons License (Attribution-NonCommercial 3.0 Unported License), as described at http://creativecommons.org/licenses/by-nc/3.0/.