Primate segmental duplication creates novel promoters for the LRRC37 gene family within the 17q21.31 inversion polymorphism region
- Cemalettin Bekpen1,2,5,
- Ibrahim Tastekin2,
- Priscillia Siswara3,
- Cezmi A. Akdis1 and
- Evan E. Eichler3,4,5
- 1Swiss Institute of Allergy and Asthma Research (SIAF), CH-7270 Davos, Switzerland;
- 2Department of Molecular Biology and Genetics, Bogazici University, Istanbul 34342, Turkey;
- 3Department of Genome Sciences, University of Washington, Seattle, Washington 98195, USA;
- 4Howard Hughes Medical Institute, Seattle, Washington 98195, USA
Abstract
The LRRC37 gene family maps to a complex region of the human genome and has been subjected to multiple rounds of segmental duplication. We investigate the expression and regulation of this gene family in multiple tissues and organisms and show a testis-specific expression of this gene family in mouse but a more ubiquitous pattern of expression among primates. Evolutionary and phylogenetic analyses support a model in which new alternative promoters have been acquired during primate evolution. We identify two promoters, Cl8 and particularly Cl3, both of which are highly active in the cerebellum and fetal brain in human and have been duplicated from a promoter region of two unrelated genes, BPTF and DND1, respectively. Two of these more broadly expressed gene family members, LRRC37A1 and A4, define the boundary of a common human inversion polymorphism mapping to chromosome 17q21.31 (the MAPT locus)—a region associated with risk for frontal temporal dementia, Parkinsonism, and intellectual disability. We propose that the regulation of the LRRC37 family occurred in a stepwise manner, acquiring foreign promoters from BPTF and DND1 via segmental duplication. This unusual evolutionary trajectory altered the regulation of the LRRC37 family, leading to increased expression in the fetal brain and cerebellum.
Gene duplication events are one of the primary sources for the evolution of novel gene functions and are, thus, important in our understanding of the evolution of species (Ohno 1970). The human genome shows a complex pattern of interspersed segmental duplication typified by a mosaic pattern of duplicons that arose recently from diverse regions of the genome. About 430 blocks of segmental duplication are identified in the human genome (Zhang et al. 2005; Bailey and Eichler 2006). Phylogenetic reconstruction has shown that the majority of recent human intrachromosomal segmental duplication blocks have formed around a set of seven core or seed duplications (Jiang et al. 2007). Interestingly, the core duplicons are highly transcribed, and numerous human/great ape gene families have recently been described that map to these regions of the genome. Several of the genes show evidence of positive selection, have radical changes in their expression profile compared with species with a single copy, and are highly copy number polymorphic (Johnson et al. 2001; Jiang et al. 2007; Han et al. 2009; Marques-Bonet and Eichler 2009). None of these gene families in human have been ascribed function, in part because of the difficulty in assaying genetic variation across these regions and distinguishing the highly identical duplicated copies.
One of these gene families, the LRRC37 (leucine-rich repeat containing 37A) family, corresponds to one of two core regions that has expanded on human chromosome 17 (Jiang et al. 2007). The putative proteins encode six internal leucine-rich repeat motifs (LRR domain). Proteins harboring leucine-rich repeats (LRRs) have been primarily implicated in functional processes related to innate immunity and neurophysiology. Toll-like receptors (TLRs) are, for example, single membrane spanning proteins whose extracellular domains are composed of LRRs, which recognize pathogen-associated molecular patterns (PAMPs) such as lipopolysaccharides (LPS), single-stranded RNA, and flagellin (Hoshino et al. 1999; Barton and Medzhitov 2002). LRR-containing proteins, such as Densin-180, Erbin, NGL, and SALM, have also been implicated in neuron-specific functions such as axonal guidance and neuronal migration and have been shown to localize to neuronal synapses (for review in detail, see Matsushima et al. 2005, 2007; Ko and Kim 2007).
Members of the LRRC37 family map to complex duplication blocks within human chromosome 17 and, in particular, define the boundary of the 970-kbp common inversion mapping to chromosome 17q21.31 (Stefansson et al. 2005; Zody et al. 2008). Inversion polymorphisms exist as two haplotypes: a direction-orientation haplotype, H1, prevalent in most human populations and an inverted haplotype, H2, which predominantly occurs in Europeans (25% allele frequency) and is associated with increased fecundity and an increase in global recombination (Evans et al. 2004; Stefansson et al. 2005). Different haplotypes of this region have been shown to be a significant risk factor locus for the tangle diseases, progressive supranuclear palsy (Baker et al. 1999), cortico-basal degeneration (Houlden et al. 2001; Pittman et al. 2004), intellectual disability (Dubourg et al. 2011), Parkinson's disease (Farrer et al. 2002; Wider et al. 2010), Alzheimer's disease (Myers et al. 2005), and predispositions to rearrangements associated with the 17q21.31 microdeletion syndrome (Sharp et al. 2006; Shaw-Smith et al. 2006; Koolen et al. 2008).
Most functional duplicate genes are associated with duplication of not only exons but most of the regulatory region of the ancestral gene (Ohno 1970). The mosaic and interspersed nature of human duplications (Bailey et al. 2002), however, allows for the potential juxtaposition of new regulatory elements from diverse regions of the genome. Relatively few examples of such innovations have been documented in human (Vandepoele et al. 2009), Drosophila (Nurminsky et al. 1998; Betran and Long 2003; Usakin et al. 2005), and Xiphophorus (Fornzler et al. 1996). Here, we analyze the evolution of the LRRC37 family and, for the first time, report that the LRRC37 family has acquired two alternative promoters from BPTF and DND1 in human. We provide evidence that these elements have been important in specifying the pattern of mRNA expression for LRRC37A1 and LRRC37A4, which map specifically to the boundary of the human-specific chromosome 17q21.31 inversion polymorphism.
Results
Promoter analysis of the LRRC37 family
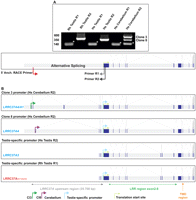
To identify the promoter initiation sites of LRRC37 genes, particularly A1 and A4, we initially compared macaque and human tissues (testis and cerebellum) using 5′ RACE (rapid amplification of cDNA ends). We cloned two distinct transcripts from human cerebellum tissue. The long transcript, Clone 3 (Cl3), is 544 bp in length and includes five novel exons of LRRC37 (specifically LRRC37A4) (Fig. 1). The shorter product, Clone 8 (Cl8), is 301 bp in length and includes three additional exons upstream of the long exon. In addition, one 347-bp transcript was cloned from human testis tissue, which is consistent with the canonical start site present in sequence databases (note: it differs by the addition of only eight additional base pairs when compared with the reference mRNA [NM_14834]). In contrast, 5′ RACE predicts two distant and distinct promoters located up to 35.7 kbp upstream of the translation initiation site (Fig. 1). Thus, in total, we identify three alternative transcription initiation sites for LRRC37A1 and A4. Analyses of the EST database and our own RT-PCR analyses show that the majority of LRRC37 transcripts initiate from the most proximal promoter identified originally from testis cDNA (T1), a testis-specific promoter region. RACE-PCR analysis confirms that this promoter at the beginning of the exon 1 region is largely testis-specific.
Identification of LRRC37 promoter regions in human and macaque. (A) RACE-PCR analysis on the cDNA isolated from the total RNA extracted from human [(Hs) Homo sapiens] and macaque [(Rh) Macaca mulatta] by using testis and cerebellum tissues. Two sets of primer pairs specific for macaque (Rh) and human (Hs) were used for final nested PCR amplification, Rh testis R1 and R2, Hs testis R1 and R2, and Hs cerebellum R1 and R2, respectively. Three PCR products were observed, cloned, and subsequently sequenced. (*) A PCR band for nonspecific amplification. (B) Schematic representation of RACE-PCR transcripts embedded on the structure of the detected LRRC37 gene structure after sequencing analysis. Promoter regions for corresponding transcripts (cerebellum clone 3 [Cl3] and cerebellum clone 8 [Cl8]) and testis-specific promoter (T1) from human (Hs Testis R2) and macaque (Rh Testis R2). (Orange arrow) The transmembrane domain (TMD).
In contrast to human, 5′ RACE only identified a short transcript of 412 bp in macaque testis corresponding to the canonical start site with no additional exons as observed in humans. RACE analysis failed to detect any transcript from macaque cerebellum likely due to the low level of expression in this particular tissue (G Giannuzzi, P Siswara, M Malig, C Bekpen, T Marques-Bonet, NISC Comparative Sequencing Center, JC Mullikin, M Ventura, EE Eichler, unpubl.). Interestingly, the corresponding putative promoter sequence for human Cl3 does not exist within the 5′ region of macaque LRRC37 genes (Fig. 2). Sequence analysis of the putative promoter region (Cl3) identified from human cerebellum revealed a 734-bp segment of shared homology (97.9% DNA sequence identity) with the promoter of another gene, dead end 1 (DND1), located on chromosome 5 in human (Fig. 2). DND1 is an evolutionarily conserved RNA binding protein that blocks the function of several miRNAs both in human and zebrafish primordial germ cells (PGCs) by binding target mRNAs and thereby restricting accessibility of miRNAs. In zebrafish, Dnd1 has been shown to be critical for the proper development of PGCs (Kedde et al. 2007). We compared available primate genomes and found that the Cl3 promoter is associated with LRRC37 genes only among humans and great apes. In all other primates, we identify a single copy of the DND1 promoter. We compared the expression profile of both the DND1 promoter and the Cl3 promoter by designing RT-PCR assays specific for corresponding transcripts. Our analysis shows that the expression of the Cl3 promoter is particularly active in fetal brain, cerebellum, liver, and testis (Supplemental Fig. S1). Notably, the DND1 transcript shows a more ubiquitous pattern of expression with highest levels in the cerebellum, thymus, lung, and testis (Supplemental Fig. S1).
Synteny map showing expressed duplicated LRRC37 genes and promoter regions. A synteny map for the LRRC37 family between mouse (mm9), macaque (rheMac2), and human (hg19) genome assembly is prepared using the Ensembl genome browser. The locations of the genes and markers are subsequently confirmed using the UCSC genome browser. The map includes only the actively transcribed LRRC37 genes (red) either reported in sequence databases or detected in our expression analysis. The locations of human LRRC37 genes on the human genome (light blue boxes) are calculated using the first exon of reported mRNA (NM_014834) for LRRC37. We have used the beginning of the LRR region to map LRRC37 genes on macaque chromosome 16 (dark green). Lrrc37 in mouse is mapped using mRNA (AK029868) for Lrrc37 in mouse chromosome 11 (cyan), and the location indicates the beginning of AK029868. The position for the promoter regions Cl3 (green), Cl8 (purple), and testis-specific (light blue) indicate the transcription start point on the respective chromosomes of the species. Arrows show the direction of a gene (5′–3′) or the transcriptional activity of the promoter region for the respective gene. Colored dotted lines connecting genes between different chromosomes indicate a possible common evolutionary history for that individual gene based on phylogenetic analysis. The black colored genes are used as syntenic markers.
Phylogenetic analysis of Cl3 promoter
To estimate the evolutionary timing of the Cl3 duplication, we analyzed the genome of available primates in conjunction with conventional PCR amplification and sequencing. We sequenced subcloned PCR products of DND1 and Cl3 promoter regions from macaque (two species and two individuals), gibbon (one individual), orangutan (four individuals), gorilla (one individual), chimp (four individuals), and human (seven individuals) (Supplemental Fig. S2). Phylogenetic analysis confirms that the DND1 promoter is found as a single copy in Old World monkey lineages and that the promoter became duplicated to the 5′-upstream region of the LRRC37 locus in the common ancestor of human and great apes. There is strong bootstrap support distinguishing the DND1 and Cl3 promoters with evidence of further sequence differentiation among humans and the African great ape Cl3 promoters. We examined the nature of the sequence differences that distinguish the Cl3 and DND1 promoters by using phylogenetic footprinting analysis (Footprinter 2.0) (Blanchette and Tompa 2003). We identified several substitutions predicted to alter transcription factor binding, which are specifically fixed in the chromosome 17 Cl3 promoter since duplication from DND1 (Supplemental Figs. S3, S4). We compared the rate of nucleotide substitution of the promoter with different intronic regions (intron 4 and intron 1). The analysis (Supplemental Fig. S5) shows that the promoter has evolved at a more rapid rate (threefold to fourfold) relative to intronic divergence.
Analysis of the Cl8 promoter
We performed a similar set of experiments with the corresponding Cl8 promoter. Comparative genome sequence analysis identified a 721-bp homologous segment (90.6% DNA sequence identity) corresponding to the promoter of the bromodomain plant homeodomain transcript factor gene (BPTF) on chromosome 17 in human. We find that the BPTF promoter exists as a single copy at the 5′-upstream region of the Bptf mRNA (NM_176850) on mouse chromosome 11. In contrast, the Cl8 promoter is duplicated to five distinct genomic locations on macaque chromosome 16. However, in orangutan, chimp, and gorilla, it is duplicated to three distinct genomic locations on chromosome 17, presumably as a result of duplications that arose after the split between the New World (NWM) and Old World monkeys (OWM) (Figs. 1, 2; Supplemental Fig. S6). In human, similar to great apes, the Cl8 promoter maps to only three locations, upstream of LRRC37A4, A3, and LRRC37B. This sequence is identified near the 5′ region of every active LRRC37 in macaque (Fig. 2). The molecular function of BPTF is unknown; however, it is shown to be expressed in embryonic and extra-embryonic tissues during early mouse embryogenesis, when it is thought to be important in trophoblast differentiation during early mouse development (Goller et al. 2008). Real-time quantitative expression analysis detects the highest activity for Cl8 in fetal brain, cerebellum, thymus, and testis in human (Supplemental Fig. S1). The Cl8 promoters show extensive sequence divergence when compared with the BPTF promoters of other mammalian species, complicating multiple sequence alignments and a more detailed phylogenetic analysis (Supplemental Figs. S6, S7).
Analysis of T1 (testis-specific) promoter
Comparative genomic analysis and BLAST search within the EST database (NCBI_dbEST) showed that almost all of the LRRC37 transcripts mapped to a promoter region at the beginning of the exon 1 region of the LRRC37 gene. We have confirmed that the promoter region at the beginning of exon 1 is active only in the testis tissues in human and macaque. The testis-specific promoter region (T1) exists at every 5′-upstream region for every active macaque and human LRRC37 gene except for LRRC375563 on macaque chromosome 16 (Fig. 2). However, our comparative genomic and dot-plot sequence alignment analysis showed no homologous DNA sequences for the exon 1 region of the human LRRC37 gene including the T1 promoter in the upstream region of the mouse Lrrc37 gene (Supplemental Fig. S8). However, in primates such as marmoset and mouse lemur (Microcebus murinus), it was possible to detect T1-like putative promoter sequences, suggesting that the T1 promoter in macaque and human arose before the anthropoid lineage diverged from more primitive lemurs and lorises (data not shown). We found that the first exon of the LRRC37 gene, which contains a putative ATG start codon, has been introduced entirely new at the 5′ region of the LRR domain of LRRC37 proteins during the expansion period in primates. Yet, the rest of the proteins seem to be conserved except for LRRC37A4 in human and LRRC376343 in macaque—both of which are missing several exons and also consist of additional splicing acceptor regions within the LRR domain (Supplemental Fig. S8).
LRRC37 genes within the boundary of H1–H2 haplotypes
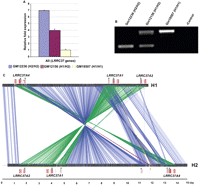
The expansion of the LRRC37 family in humans is associated with a 970-kbp inversion polymorphism that occurs as two haplotypes: H1 (direct orientation) and H2 (reverse orientation) (Fig. 3; Supplemental Figs. S9, S10). Previously, we showed that the breakpoints of the inversion map specifically within the LRRC37 genes LRRC37A1 and A4. We compared the sequence of the two haplotypes in the context of these putative promoters and identified a second Cl8 promoter located upstream of LRRC37A4 specifically for the H2 haplotype (Fig. 3). In addition, the normal Cl8 promoter maps significantly further upstream (160.740 bp) when compared with H1, which is only 34.042 bp upstream of the long coding exon. To assess the potential impact of these regulatory changes, we assayed the expression of the LRRC37 family from lymphoblastoid cell lines derived from homozygous and heterozygous genotypes. We find that the H2 haplotype shows at least a sevenfold increase in expression of LRRC37 genes when compared with the H1 haplotypes (Fig. 3). Due to technical difficulties arising from alternative splicing, we were unable to perform real-time PCR specifically for the Cl3-derived transcript. Thus, this differential regulation of LRRC37 genes between H1 and H2 haplotypes may be explained by the chromosomal rearrangements due to the inversion polymorphism or alternatively due to other sequence differences that have occurred on the H2 lineage. Additionally, to find other putative duplicated sequences similar to Cl8 and Cl3 promoters within the 17q21.31 inversion polymorphism for both H1 (1.169 kbp) and H2 (1.481 kbp) haplotypes that are deeply sequenced and assembled by Zody et al. (2008), we have applied the DupMasker program developed by Jiang et al. (2008). DupMasker analysis showed that there are 527 different duplicons for the H1 haplotype, whereas for the H2 haplotype, the numbers of the duplications were 688. Although there are duplicated genomic segments from different chromosomal locations, we have found that most of the duplications within the H1 and H2 regions were originating from different genomic locations within chromosome 17 (Supplemental Tables 1, 2).
Structural polymorphism of LRRC37 genes as H1 and H2 haplotypes. (A) Relative fold expression of the LRRC37 family was detected by real-time PCR using the lymphocyte-derived cell lines. The figure shows the relative fold expression of all LRRC37 genes (All) between GM12236 (H2/H2), GM12156 (H1/H2), and GM18507 (H1/H1) individuals. Expression data were normalized against the housekeeping gene UBE1. (B) PCR analysis is performed to confirm the genotype of H1 and H2 individuals using a diagnostic indel (Evans et al. 2004). (C) A mirorepeats alignment comparing the human chromosome 17 H1 reference sequence to a sequence from an alternate H2 haplotype. The alignment depicts a 1.5-Mb inversion region of the MAPT locus. (Arrow) The transcription start point from Cl3 (green), Cl8 (purple), and testis-specific (light blue) promoters at the 5′ upstream region of LRRC37 genes LRRC37A1, A2, and A4, respectively. (Red lines) The exons for the respective LRRC37 genes. (Green lines) The boundary of the inversion region within the LRRC37 genes. Each line (green or blue) shows 100 bp of homology. The figure is drawn to scale.
Luciferase activity of LRRC37 promoters
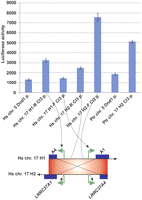
We analyzed the activity of specific Cl3 promoters using a luciferase reporter assay. We find that all promoters, with the exception of the H1-F Cl3 promoter located on the Chr17q21.31 inversion region, show higher luciferase activity in human HEK293FT cells when compared with the original DND1 promoter located on Chr5 (Fig. 4). A similar result is observed with constructs derived from chimpanzee—i.e., the duplicated copy of the Ptr-H2 promoter consistently shows higher activity than the ancestral Ptr-DND1 promoter (Fig. 4). Confirming our previous real-time quantitative PCR analysis (Fig. 3), we find that Cl3 promoters from the H2 haplotype show higher luciferase activity when compared with Cl3 promoters derived from the H1 haplotype. Interestingly, we find that the Cl3 promoter corresponding to H2 LRRC37A4 shows the highest levels of promoter activity (eightfold greater activity when compared with the ancestral DND1 human promoter region). These data suggest that sequence differences within Cl3 have been critical in altering the expression of this copy in the cerebellum, perhaps as a result of positive Darwinian selection (Stefansson et al. 2005).
Luciferase activity assay for promoter regions of the LRRC37 family. (Top panel) Luciferase activity of the ancestral copy of the DND1 promoter region from the (Hs) human (H. sapiens) and (Ptr) chimpanzee (Pan troglodytes) DND1 promoters and a duplicated copy of the promoter regions from the human Chr 17q21.31 H1 haplotype (Hs Chr17 H1-R Cl3 p and Hs Chr17 H1-R Cl3 p), H2 haplotype (Hs Chr17 H2-R Cl3 p and Hs Chr17 H2-R Cl3 p), and PTR chimpanzee (Chr 17 H2 Cl3 p) in HEK293FT cells. (Bottom panel) Schematic representation of H1 and H2 haplotypes on the human genome.
Discussion
There are 13 copies of the LRRC37 family distributed along human chromosome 17. This is in contrast to mouse and most other mammalian genomes where only a single copy can be identified (Supplemental Fig. S9). Evolutionary and phylogenetic analysis indicates a specific expansion of the LRRC37 family within the anthropoids with continued expansion and significant restructuring of this locus in humans and great apes. The sequence has been classified as a “core” duplicon because it defines one of two focal points for the accretion of complex segmental duplications on human chromosome 17 (Jiang et al. 2007). We currently estimate that approximately half of the gene family has either been pseudogenized or shows no evidence of transcription (G Giannuzzi, P Siswara, M Malig, C Bekpen, T Marques-Bonet, NISC Comparative Sequencing Center, JC Mullikin, M Ventura, EE Eichler, unpubl.; this study).
The LRRC37 core duplicon has been implicated in genomic instability associated with disease. For example, one member, LRRC37B, maps at or close to the breakpoints associated with the NF1 microdeletion syndrome (Bengesser et al. 2010). Two other members of the LRRC37 family, LRRC37A1 and A4, define the breakpoints of the common MAPT inversion polymorphism at chromosome 17q21.31 (Zody et al. 2008). The H1 and H2 haplotypes have been the focus of both potential positive selection within the human species (Stefansson et al. 2005) as well as risk factors for several diseases including intellectual disability (Hardy et al. 2005; Dubourg et al. 2011), Alzheimer's disease, and Parkinsonism (Farrer et al. 2002; Myers et al. 2005; Wider et al. 2010).
In this context, we find it particularly noteworthy that other genes thought to encode LRR-containing proteins have been implicated in neuropsychiatric disorders (Matsushima et al. 2005), epilepsy (Kalachikov et al. 2002) and Alzheimer's (Majercak et al. 2006). Using lymphoblastoid cell lines, we show that individuals who carry the H2 haplotype express LRRC37 at a sevenfold higher level than H1 homozygotes, with heterozygotes showing an intermediate pattern of expression. These findings are experimentally validated based on our luciferase assays, which suggest that changes in the nature of the promoter itself may have been sufficient to drive this up-regulation. A previous study indicated a reduced level of brain expression for most other genes on the H2 haplotype (Myers et al. 2007) when compared with H1. Our results suggest that LRRC37 is unusual, being one of the only genes up-regulated on the H2 haplotype.
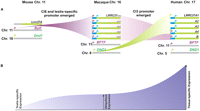
Our comparative analyses suggest that the expression of the LRRC37 family evolved from testis-specific pattern expression (as seen in mouse) to a more ubiquitous expression in macaque and human (Figs. 1, 2). We propose that these expression changes were coordinated with increased segmental duplication copy number and duplicative transposition of regulatory elements from other genes. In the common ancestor with New World monkeys, the “BPTF” promoter duplicated to the 5′-upstream region of the LRRC37 family. This BPTF-derived promoter subsequently spread to the 5′ region of almost every active LRRC37, as indicated by the macaque genome assembly. It is interesting that in humans the BPTF duplicated promoter is restricted to only two copies: LRRC37A4 and LRRC37B. LRRC37B is indicated to be one of the hominoid-specific genes that is up-regulated specifically in human prefrontal cortex (PFC) (Zhang et al. 2011). Furthermore, during the evolution of human and great apes, the “DND1” promoter duplicatively transposed to the 5′-upstream region of LRRC37A1 and A4. These genes demarcate the boundary of the common inversion polymorphism. Our data suggest that these segmental duplications have been important in broadening the expression profile of the LRRC37 family, especially in increasing expression in the cerebellum. Overall expression of the LRRC37 family in human is enriched at least 30-fold in thymus and cerebellum (G Giannuzzi, P Siswara, M Malig, C Bekpen, T Marques-Bonet, NISC Comparative Sequencing Center, JC Mullikin, M Ventura, EE Eichler, unpubl.).
The fate of duplicated genes is complex, with most becoming pseudogenized during the course of evolution because of a relaxation of functional constraint. Radical shifts in expression profiles of duplicated genes are consistent with neofunctionalization (Conant and Wolfe 2008). In most cases in which the duplicate retains function, the regulatory region in the ancestral gene is also transferred to the daughter gene. However, in rare cases, new duplicates can acquire a promoter from another gene as has been shown in Drosophila (Nurminsky et al. 1998; Betran and Long 2003; Usakin et al. 2005), Xiphophorus (Fornzler et al. 1996), and human (Vandepoele et al. 2009). Our analysis of the LRRC37 family suggests that it has acquired two promoters from other genes in a stepwise manner during 40 million years of primate evolution (Fig. 5). The mosaic and interspersed nature of primate duplications provides an extraordinary milieu for such innovations in transcription to emerge. Our results demonstrate the speed at which expression changes can occur for genes embedded within segmental duplications. The functional implications of these changes as they relate to the specific tissues (such as the cerebellum or fetal brain) still await future investigation.
Evolution at the regulatory region of the LRRC37 family. A model for the evolution of promoter regions of LRRC37 is depicted. (A) The LRRC37 family acquired alternative promoter regions, first, from the BPTF promoter region within the macaque lineage; and, second, from the DND1 promoter after the split between New World and Old World monkeys, respectively. An additional promoter, which is amplified from human testis and macaque testis tissues, is detected just upstream of the predicted long coding exon containing the ATG start codon in macaque and human. (B) A schematic representation of the regulation of gene expression within the LRRC37 family. The LRRC37 family evolved from testis-specific expression in mouse to ubiquitous or tissue-specific expression, such as cerebellum and thymus, in human.
Methods
RT-PCR
cDNA was prepared using the RT-PCR kit (Roche) according to the manufacturer's instructions. Total RNA used for cDNA preparation was extracted from total RNA (RNeasy; QIAGEN) of Rh (macaque) tissues, and the total RNA for Hs (human) tissues was purchased from Clontech. UBE1 was used as positive control. PCR was performed in 20-μL reactions composed of 0.8 μL of a 10 μM dilution of the forward primer and reverse primer and 10 μL of Roche (11636103001) PCR Master Mix. Please see Supplemental Table 3 for the PCR conditions and primers for cloning the promoter constructs for Cl3 and DND1.
Real-time PCR
Various LRRC37 transcripts were detected by a quantitative PCR assay using the LightCycler SYBR Green System (Roche) with exon 6 and 8 primers. cDNA was synthesized using mRNA prepared from lymphoblast cell lines. The amount of measured transcripts was normalized to the amount of the GAPDH and UBE1 transcripts. Please see Supplemental Table 3 for the real-time PCR conditions and primers for cloning promoter constructs for Cl3, Cl8, and DND1.
5′ RACE-PCR
Single-stranded cDNA was prepared using a rapid PCR purification kit (Roche). The terminal deoxynucleotidyl transferase reaction was prepared as follows: 16.5 μL of cDNA, 5 μL of TdT + Reaction buffer (Amersham), and 2.5 μL of dCTP (2 mM) were incubated for 3 min at 94°C; then 1 μL of TdT was added and incubated for 15 min at 37°C, followed by an inactivation step for 5 min at 65°C. PCR was performed on the cDNA + poly(C) using the primer 5′Anc and LRRC37R. PCR products were purified using the rapid PCR purification kit, and a second round of nested PCR was performed using the primers UAP and LRRC37 R1 and R2 for human and macaque (please see Supplemental Table 3). The 600-bp, 350-bp, and 140-bp PCR products were cloned to PGEM-T Easy, and insert sequences were determined by sequencing.
Phylogenetic analyses
All multiple sequence alignments were generated using ClustalW (Thompson et al. 1994; Chenna et al. 2003). We constructed neighbor-joining phylogenetic trees (MEGA 3.1) (Kumar et al. 1994).
Cloning of promoter constructs
XhoI–HindIII fragments were amplified from the original Dnd1 promoter located on chromosome 5 and each of the duplicated H1 and H2 versions of the Cl3 promoters using the degenerate primers (Supplemental Table 3). The same primers were used to amplify the original DND1 promoter and duplicated H1 and H2 promoters from the chimpanzee genome, making use of the homologous regions between human and chimpanzee. Each of these fragments was ligated into the XhoI–HindIII-digested pGL3-Basic vector to allow transcription of the firefly luciferase gene under the control of these fragments (Promega). As a negative control, reverse-orientation versions of each of the fragments described above were cloned (Supplemental Table 3). Each of these reverse-oriented fragments was ligated into the XhoI–HindIII-digested pGL3-Basic vector to allow transcription of the firefly luciferase gene under the control of these fragments (Promega). All of the resultant vectors were verified by restriction analysis and sequencing.
Cell culture and transfection
HEK293FT cells were grown in DMEM (Life Technologies; GIBCO) supplemented with 10% (v/v) heat-treated FBS, 2 mM glutamine (Life Technologies; GIBCO), and 100 U of penicillin–streptomycin (Life Technologies; GIBCO). The culture was incubated at 37°C including 5% CO2. For luciferase activity assays, cells were seeded on six-well plates (∼2 × 105 cells/well) and incubated overnight. Then the cells were cotransfected with the plasmids described above together with beta-gal control plasmid (pcDNA4) using the FuGENE HD transfection reagent (Roche) as described by the manufacturer. Luciferase activity was performed by using the dual luciferase assay (Promega) according to the manufacturer's recommendations, normalized against beta-gal (Promega) activity for respective transfection.
Data access
Cloned mRNA fragments of 5′-UTR regions from LRRC37A1 and A4 genes used in this work have been submitted to the NCBI GenBank (http://www.ncbi.nlm.nih.gov/genbank/) under accession numbers JQ358760, JQ358761, JQ358762, and JQ358763.
Acknowledgments
We thank Lin Chen for computational support, Guiliana Giannuzzi for access to unpublished data, and Tonia Brown for her help during the preparation of this manuscript. We also thank Stefan Fuss, Arzu Celik, and Ibrahim Yaman for their technical support. Cemalettin Bekpen is supported by the project FP-7 Regpot 2009-1 MBG Bridge. This work was supported, in part, by NIH grant GM058815 to E.E.E. E.E.E. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
-
↵5 Corresponding authors.
E-mail cemalettin.bekpen{at}boun.edu.tr.
E-mail eee{at}gs.washington.edu.
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.134098.111.
- Received October 28, 2011.
- Accepted March 7, 2012.
This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date (see http://genome.cshlp.org/site/misc/terms.xhtml). After six months, it is available under a Creative Commons License (Attribution-NonCommercial 3.0 Unported License), as described at http://creativecommons.org/licenses/by-nc/3.0/.