Locus- and domain-dependent control of DNA methylation at mouse B1 retrotransposons during male germ cell development
- Kenji Ichiyanagi1,9,
- Yungfeng Li1,
- Toshiaki Watanabe2,
- Tomoko Ichiyanagi1,
- Kei Fukuda1,3,
- Junko Kitayama4,
- Yasuhiro Yamamoto1,
- Satomi Kuramochi-Miyagawa5,
- Toru Nakano5,
- Yukihiro Yabuta6,7,
- Yoshiyuki Seki7,8,
- Mitinori Saitou6,7 and
- Hiroyuki Sasaki1
- 1Division of Epigenomics, Medical Institute of Bioregulation, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan;
- 2Department of Cell Biology, School of Medicine, Yale University, New Haven, Connecticut 06510, USA;
- 3Department of Genetics, School of Life Science, The Graduate University for Advanced Studies (SOKENDAI), Mishima, Shizuoka 411-8540, Japan;
- 4Division of Agricultural Genetics, National Institute of Genetics, Mishima, Sizuoka 411-8540, Japan;
- 5Department of Pathology, Graduate School of Frontier Biosciences, Osaka University, Suita, Osaka 565-0871, Japan;
- 6Department of Anatomy and Cell Biology, Graduate School of Medicine, Kyoto University, Kyoto 606-8501, Japan;
- 7Laboratory for Mammalian Germ Cell Biology, Center for Developmental Biology, RIKEN Kobe Institute, Kobe, Hyogo 650-0047, Japan;
- 8Department of Bioscience, School of Science and Technology, Kwansei Gakuin University, Sanda, Hyogo 669-1337, Japan
Abstract
In mammals, germ cells undergo striking dynamic changes in DNA methylation during their development. However, the dynamics and mode of methylation are poorly understood for short interspersed elements (SINEs) dispersed throughout the genome. We investigated the DNA methylation status of mouse B1 SINEs in male germ cells at different developmental stages. B1 elements showed a large locus-to-locus variation in methylation; loci close to RNA polymerase II promoters were hypomethylated, while most others were hypermethylated. Interestingly, a mutation that eliminates Piwi-interacting RNAs (piRNAs), which are involved in methylation of long interspersed elements (LINEs), did not affect the level of B1 methylation, implying a piRNA-independent mechanism. Methylation at B1 loci in SINE-poor genomic domains showed a higher dependency on the de novo DNA methyltransferase DNMT3A but not on DNMT3B, suggesting that DNMT3A plays a major role in methylation of these domains. We also found that many genes specifically expressed in the testis possess B1 elements in their promoters, suggesting the involvement of B1 methylation in transcriptional regulation. Taken altogether, our results not only reveal the dynamics and mode of SINE methylation but also suggest how the DNA methylation profile is created in the germline by a pair of DNA methyltransferases.
In mammals, up to ∼50% of the genome comprises repetitive sequences (Lander et al. 2001; Waterston et al. 2002; Lindblad-Toh et al. 2005; Mikkelsen et al. 2005, 2007; Warren et al. 2008; Elsik et al. 2009), including long interspersed elements (LINEs), short interspersed elements (SINEs), and long terminal repeat (LTR) retrotransposons (Deininger and Batzer 2002; Deininger et al. 2003). The activities of LINEs and LTR retrotransposons, which can act as aggressive mutators, are suppressed by epigenetic mechanisms, such as DNA methylation, in both somatic and germ cells (Walsh et al. 1998; Bourc'his and Bestor 2004; Tsumura et al. 2006). DNA methylation at CG dinucleotides is a key epigenetic modification in mammals (Bird 2002) that is essentially heritable in somatic cells. In contrast, it is modifiable in early embryos and germ cells. Primordial germ cells (PGCs) undergo extensive erasure of methylation (Hajkova et al. 2002; Seki et al. 2005), and in males, prospermatogonia (gonocytes) acquire new methylation marks (Sasaki and Matsui 2008) (for stages during male germ cell development, see Fig. 1A). The methylation levels of LINEs and LTR retrotransposons follow these global changes in male germ cell development (Lane et al. 2003; Lees-Murdock et al. 2003; Kato et al. 2007; Popp et al. 2010).
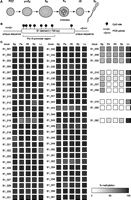
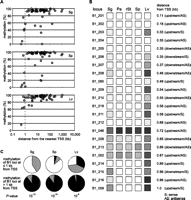
DNA methylation at B1 loci in germ and somatic cells. (A) Schematic representation of male germ cell development. (PGC) Primordial germ cell; (proSg) prospermatogonium; (Sg) spermatogonium; (Pa) pachytene spermatocyte; (rSt) round spermatid; (Sp) spermatozoon. (B) Experimental protocol for analysis of individual B1 elements. B1 elements (open rectangle) containing six to eight CpG sites (shown as lollipops) were selected, and primers (gray arrows) were designed for unique flanking regions. (C) Methylation of individual B1 elements. Methylation levels in the indicated cells (noted above the columns) at B1 loci designated on the left (B1_00–B1_062) are represented as filled squares on the basis of a grayscale: from white = 0% methylation to black = 100% methylation. (Lv) Liver cells.
SINEs are 100–500 bp in length and have sequence features that are distinct from those of LINEs and LTR retrotransposons. For example, SINEs possess an RNA polymerase III (Pol III) promoter (Kramerov and Vassetzky 2005), whereas LINEs and LTR retrotransposons possess a RNA polymerase II (Pol II) promoter. The mouse genome contains about 1.5 million SINE copies, which together constitute 8% of this genome (Waterston et al. 2002). Mouse SINEs are classified into families of B1, B2, B3, B4, MIR, and ID. B1 and B2, the largest families with 570,000 and 380,000 copies, respectively, contain currently active copies.
B1 is an ∼150-bp element in rodents that shares a common origin with Alu SINEs in primates. Interestingly, although B1 and Alu SINEs have independent evolutionary histories, they are both accumulated in similar gene-rich regions of the respective genomes, suggesting their roles in gene regulation (Lander et al. 2001; Waterston et al. 2002; Polak and Domany 2006; Tsirigos and Rigoutsos 2009). Analyses of methylation of the bulk of B1 copies have revealed that B1 elements are hypomethylated in PGCs and hypermethylated in late prospermatogonia, spermatozoa, and somatic cells (Kato et al. 2007; Meissner et al. 2008; Popp et al. 2010).
Mammals have two DNA methyltransferases, DNMT3A and DNMT3B, that are responsible for de novo methylation and a regulatory protein with no catalytic activity, DNMT3L (Bestor 2000). Mutational studies in mice revealed that the two enzymes have redundant activities for de novo methylation of L1 (a member of LINEs) and intracisternal A particle (IAP, a member of LTR retrotransposons), whereas DNMT3L is critical for de novo methylation of both elements (Walsh et al. 1998; Bourc'his and Bestor 2004; Kato et al. 2007). Methylation of L1 and IAP has been recently proposed to be guided by a class of germ cell–specific small RNAs, designated Piwi-interacting RNAs (piRNAs), which are produced by the actions of PIWIL2 (also called MILI), PIWIL4 (MIWI2), DDX4 (MVH), MOV10L1, and PLD6 (MITOPLD) (Aravin et al. 2007, 2008; Carmell et al. 2007; Kuramochi-Miyagawa et al. 2008, 2010; Zheng et al. 2010; Watanabe et al. 2011a). Tudor-domain proteins, TDRD1 and TDRD9, and GASZ are also involved in piRNA biogenesis and de novo L1 methylation in male germ cells (Ma et al. 2009; Reuter et al. 2009; Shoji et al. 2009). The piRNA-dependent DNA methylation in mammals is analogous to the RNA-directed sequence-specific DNA methylation in higher plants (Matzke et al. 2007). However, the generality of the piRNA-dependent DNA methylation remains unclear because only a few retrotransposons have been analyzed until this date.
Here we analyzed the DNA methylation status of a number of individual B1 loci in male germ cells at various developmental stages as well as in spermatogonia deficient in de novo methylation or piRNA biogenesis. Our results revealed that most loci were stably hypermethylated during spermatogenesis, with some variations dependent on the locus and domain. By incorporating these results from both wild-type and mutant germ cells, we propose a model that explains how de novo methylation of B1 is regulated in the germ cell genome. Moreover, we provide data that suggest a role for B1 in transcriptional regulation of testis-specific genes and discuss the impact of B1 retrotransposition in rodent evolution.
Results
Locus-dependent variations in B1 methylation in male germ cells
The consensus sequence of young B1 subfamilies has eight CG sites, most of which reside in the Pol III promoter region (Fig. 1B). Previous studies (Kato et al. 2007; Popp et al. 2010) have reported on the DNA methylation status of the bulk of genomic B1 copies in some somatic tissues and germ cells, providing rough estimates of their overall methylation levels. However, the methylation levels were most likely underestimated because of the occurrence of frequent CG-to-TG mutations (unmethylated CG and TG are indistinguishable in bisulfite sequencing readouts) (see legend for Supplemental Fig. S1). More importantly, the extent of variation in methylation levels among individual loci remains unknown.
We analyzed 62 individual B1 elements that were selected on the basis of their size, sequence conservation, and number of CG sites (Fig. 1C; see Methods for the detailed selection method). In liver cells (a somatic control), almost all loci examined were uniformly hypermethylated with an average methylation level of 92%, with only one exception (B1_060; 16% methylation). In spermatogenic cells, most loci (n = 52) showed stable hypermethylation (>80% methylation), while 10 loci showed lower levels of methylation. Of these 10 loci, three (B1_005, B1_006, and B1_052) showed intermediate levels of methylation (40%–70%), and three others (B1_019, B1_060, and B1_062) showed very low or virtually no methylation throughout spermatogenesis. The remaining four (B1_015, B1_028, B1_037, and B1_059) showed intermediate levels of methylation in spermatogonia and became hypomethylated during (spermatocytes) and after meiosis (spermatids and spermatozoa). Taken together, male germ cells, particularly those at later spermatogenic stages, exhibited significantly less methylation at some B1 loci compared with liver cells.
The existence of some hypomethylated loci in male germ cells prompted us to examine the expression levels of B1 RNA in the testis. Consistent with a previous study (Li et al. 1999), B1 RNA of ∼150 nt was accumulated in the testis but not in other tissues such as the brain, heart, lung, intestine, liver, or kidney (Supplemental Fig. S2A). We infer that hypomethylated loci may be the source of testis-accumulated B1 RNA, but a high similarity in nucleotide sequence among young B1 elements precluded detailed analysis of locus-specific B1 transcription. We also found that B1 elements were poorly methylated in oocytes (Supplemental Fig. S1), wherein B1 expression is very high (Kaplan et al. 1985; Peaston et al. 2004). These results are consistent with the inverse correlation between DNA methylation and Pol III promoter activity.
B1 methylation is independent of piRNA production
We next analyzed the methylation status of the bulk of B1 elements in male germ cells (Supplemental Fig. S1). These results suggest that de novo methylation of B1 occurred between embryonic day 12.5 (E12.5; PGCs) and postnatal day 0 (P0; prospermatogonia). Because de novo methylation at L1 and IAP involves prospermatogonial piRNAs (Aravin et al. 2007, 2008; Kuramochi-Miyagawa et al. 2008, 2010; Watanabe et al. 2011a), we analyzed the previously published expression profiles of small RNAs (24–31 nt in length) in E16.5 (fetal) and P10 (prepuberal) testes (Aravin et al. 2007, 2008).
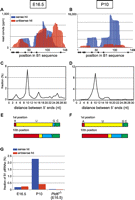
B1-derived piRNAs corresponding to various portions of the B1 sequence were found in the E16.5 library (Fig. 2A). Importantly, a substantial fraction of the sense and antisense pairs had a 10-bp complementary sequence (Fig. 2C), and the majority had uracil at the first position and adenine at the tenth position (Fig. 2E); these are the signatures of piRNAs produced via the ping-pong cycle (Brennecke et al. 2007; Gunawardane et al. 2007). In P10 testes, where spermatogonia are the predominant cell type, the number of B1-derived sense piRNAs increased by >10-fold, whereas the number of antisense piRNAs increased only slightly (Fig. 2B,G). These piRNAs also showed the signatures of ping-pong cycle products (Fig. 2D,F).
Analysis of B1-derived small RNAs in fetal and prepuberal testes. Small RNAs in E16.5 (Aravin et al. 2008) (ACE) and P10 (Aravin et al. 2007) (BDF) testes were mapped to B1_Mus1 consensus sequences. (A,B) Relative read counts (number of hits divided by the total sequence reads, expressed as parts per million, ppm) of sense (blue) and antisense (red) small RNAs. (C,D) The start position of each sense-strand read was compared with that of all antisense-strand reads. The percentages of relative length overlap between sense and antisense reads are shown. (E,F) Nucleotide compositions at the first and tenth positions of B1-derived small RNAs. (G) The relative abundance of B1-derived small RNAs. The fractions of sense (blue) and antisense (red) reads in the total sequence reads are shown. The data for piRNAs in Pld6/MitoPLD KO prospermatogonia were obtained from Watanabe et al. (2011a).
The presence of B1-derived piRNAs in prospermatogonia raised the possibility of these piRNAs playing a role in de novo methylation of B1 elements, similar to that in de novo methylation of L1 and IAP elements. Therefore, we analyzed the B1 methylation status in spermatogonia from Pld6 knockout (KO) mice in whom prospermatogonial piRNAs were markedly decreased (Fig. 2G; Watanabe et al. 2011a). The methylation levels at the B1 loci analyzed were largely unaffected (Fig. 3A,E). Further analysis on selected B1 loci in wild-type, Pld6 KO, and Piwil2 KO prospermatogonia revealed no significant decrease in B1 methylation in these mutant cells (Supplemental Fig. S3). These results indicate that B1 elements could be de novo methylated in prospermatogonia independent of the piRNA pathway. Moreover, the accumulation of full-length B1 RNA in spermatogonia was not increased by the Pld6 or Piwil2 KO mutation (Supplemental Fig. S2B). Thus, B1-derived piRNAs have little effect on DNA methylation and RNA stability of B1, although it remains possible that these piRNAs play a role in post-transcriptional silencing of mRNAs that contain a B1 sequence, because DICER-dependent endogenous siRNAs of B1 origin regulate such mRNAs in preimplantation embryos (Ohnishi et al. 2011).
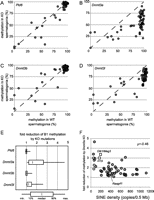
Effects of mutations in piRNA biogenesis and DNA methyltransferases on de novo B1 methylation. Methylation levels (y-axis) at individual B1 loci in spermatogonia of Pld6−/− (A), Dnmt3a2lox/1lox TNAP-Cre (B), Dnmt3b2lox/1lox TNAP-Cre (C), and Dnmt3l−/− (D) are plotted against their methylation levels in wild-type spermatogonia (x-axis). Dashed lines show the y = x slope. (E) Fold reductions in individual B1 methylation due to mutations. Only the data for B1 loci with ≥80% methylation in wild-type spermatogonia are included. (F) Fold reductions in methylation due to Dnmt3a KO mutation at individual B1 loci (dark gray circles) are plotted against the SINE density in the flanking genomic regions (0.25 Mb of each side). B1 loci selected from SINE-poor domains (B1_101–B1_105) are shown as light gray circles. The methylation levels at three paternally methylated regions for genomic imprinting (H19, Dlk1/Meg3, and Rasgrf1) in wild-type and Dnmt3a KO spermatogonia were examined as described previously (Kato et al. 2007), and the results are shown as open squares. Spearman's correlation coefficient (ρ) is shown on the right.
We confirmed that methylation of the bulk of Gf and A monomers of L1 was markedly affected in the Pld6 KO spermatogonia (Supplemental Fig. S4). In stark contrast, methylation of the bulk of IAP and MMERVK10C elements, whose piRNAs are abundant in prospermatogonia (Kuramochi-Miyagawa et al. 2008), was largely unaffected (Supplemental Fig. S4), although a previous report showed that specific copies of an IAP subtype were affected in piRNA pathway mutants (Kuramochi-Miyagawa et al. 2008, 2010; Watanabe et al. 2011a). Thus, even for autonomous retrotransposons, different elements show different levels of piRNA dependency for targeted DNA methylation.
Dependence of B1 methylation on Dnmt3a, Dnmt3b, and Dnmt3l
To understand which DNMT3 family proteins were involved in de novo methylation of B1, we examined methylation levels at B1 loci in spermatogonia from conditional KO mutants for Dnmt3a (Dnmt3a2lox/1lox, TNAP-Cre) and Dnmt3b (Dnmt3b2lox/1lox, TNAP-Cre), as well as those from conventional Dnmt3l KO mice. In conditional KO mice, Dnmt3a and Dnmt3b are inactivated in germ cells before the prospermatogonial stage (Kaneda et al. 2004; Kato et al. 2007).
At most B1 loci examined, the methylation level was substantially reduced in Dnmt3a KO spermatogonia (Fig. 3B,E) but not in Dnmt3b KO spermatogonia (Fig. 3C,E). Dnmt3l KO spermatogonia showed methylation that was reduced to an intermediate level (Fig. 3D,E). These results are consistent with the fact that DNMT3A cooperates with DNMT3L for de novo methylation in germ cells, but the milder defect in Dnmt3l KO spermatogonia suggests that DNMT3A also acts independent of DNMT3L. This is in contrast with previous findings on IAP and L1 elements, where Dnmt3l KO severely affected methylation but single Dnmt3a or Dnmt3b KO did not (Bourc'his and Bestor 2004; Kato et al. 2007).
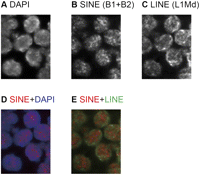
Of note, the effect of Dnmt3a KO on methylation varied among the B1 loci, ranging from almost no change to about a fivefold reduction (Fig. 3E). This suggests that although Dnmt3b KO did not affect B1 methylation, Dnmt3b may have a back-up role. We explored the genomic features that affected the dependence on Dnmt3a and found that methylation tended to be more Dnmt3a-dependent if the B1 locus was in a SINE-poor, LINE-rich, gene-poor, or low G+C genomic region (Fig. 3F; Supplemental Fig. S5). Among these features, SINE density showed the highest correlation to Dnmt3a dependency (Spearman's ρ = −0.46, +0.39, −0.27, and −0.17 for Dnmt3a dependency vs. SINE density, LINE density, gene density, and G+C content, respectively). To confirm this correlation, we selected an additional five B1 loci (B1_101–B1_105) that were present in SINE-poor regions (<100 SINEs/0.5 Mb) and examined their methylation levels in wild-type and Dnmt3a KO spermatogonia. These loci indeed showed high levels of Dnmt3a dependency (Fig. 3F). Interestingly, three paternally methylated regions from the imprinted loci also followed the same rules (Fig. 3F). De novo methylation of the H19 and Dlk1/Meg3 differentially methylated regions (DMRs) were highly dependent on Dnmt3a; these DMRs were located in SINE-poor regions. De novo methylation of the Rasgrf1 DMR did not depend on Dnmt3a; this DMR was located outside the SINE-poor regions. Interestingly, SINE-poor domains were not distributed evenly within the nucleus; fluorescence in situ hybridization (FISH) using SINE (B1 plus B2) and LINE (L1) probes showed that SINE-poor domains tended to be located at the periphery of prospermatogonial nuclei (Fig. 4; Supplemental Fig. S6). These domain-dependent distribution patterns might be involved in the observed variations in Dnmt3a dependency (see Discussion).
Spatial distributions of SINE and LINE elements in prospermatogonial nuclei. Frozen sections of E15.5 testis were stained with 4′,6-diamidino-2-phenylindole (DAPI) (A) and hybridized against Cy3-dUTP-labeled B1/B2 probes (B) and green dUTP-labeled L1 probes (C). (D) Merged image of panels A and B. (Red) SINE; (blue) DAPI. (E) Merged image of panels B and C. (Red) SINE; (green) LINE. Images of non-adhering germ cells in a seminiferous tubule are shown (600× magnification).
B1 loci in Pol II promoter regions are protected from methylation
As described above, B1 loci showed large differences in methylation in wild-type germ cells despite the fact that the internal nucleotide sequences did not significantly differ between the hyper- and hypomethylated loci (Supplemental Fig. S7). We explored what genomic features might be associated with the hypomethylated loci. The local LINE, SINE, and gene densities, G+C content, and the location in terms of the Pol II transcription unit (i.e., genic vs. intergenic) were not major factors contributing to protection from methylation (Supplemental Fig. S8A–E). However, we found that B1 loci located at <1 kb from a transcription start site (TSS) were generally hypomethylated, regardless of their orientation relative to the transcript (Fig. 5A). The CpG density of the neighboring regions was also correlated (Supplemental Fig. S8F), which may have reflected the CpG-rich nature of the regions around TSSs.
DNA methylation levels and proximity to TSSs. (A) Methylation levels at individual B1 loci (y-axis) are plotted against their distances to the nearest TSS (x-axis). Results for spermatogonia, spermatozoa, and liver are shown. Results for pachytene spermatocytes and round spermatids were nearly the same as those for spermatozoa (see Fig. 1C). (B) Methylation levels at B1 loci (B1_201–B1_216) that are present at <1 kb from TSS are shown on a grayscale as in Figure 1C. TSS-proximal B1 loci in the initial group (B1_060, B1_019, B1_046, B1_028, B1_052, and B1_059) are also included. (C) Statistics for B1 methylation and their distances from TSS. The 78 B1 loci (62 + 16) are categorized into those at ≤1 kb from TSS (upper pie charts) and those at >1 kb (lower pie charts). The loci are further categorized into 0%–19% methylation (white), 20%–79% methylation (gray), and 80%–100% methylation (black). The P-values from χ2 tests are shown at the bottom.
To confirm this correlation, we randomly selected an additional 16 B1 loci (B1_201–B1_216) located within 1 kb from TSS and analyzed their methylation levels in spermatogenic cells. Most of these loci were hypomethylated in spermatogenic cells (Fig. 5B). It is noteworthy that some loci (B1_203, B1_206, B1_208, B1_209, B1_211, B1_028, B1_215, B1_216, and B1_059) were moderately (17%–58%) methylated in spermatogonia and then lost methylation in pachytene spermatocytes. Overall, promoter-proximal B1 loci were significantly less methylated than other loci (p < 10−8) (Fig. 5C). This trend was more evident in germ cells because B1s located at >0.3 kb away from TSSs showed denser methylation in liver cells than in germ cells (Fig. 5B). The higher methylation levels at these B1 loci in liver cells were not correlated with the methylation levels at the TSS regions themselves because these regions were consistently unmethylated in all cells examined (Supplemental Fig. S9).
In general, the density of retrotransposons is extremely low in Pol II promoter regions (Fig. 6A). Although this is also true for B1 elements, we found that they were at a higher density in testis-specific promoters compared with all promoters (Fig. 6A,B; p < 10−16 by χ2 test). These results suggest the possibility that the presence of a B1 element in the proximity of TSS is associated with suppression of genes in somatic cells. Furthermore, differential methylation of TSS-proximal B1 elements between germ and somatic cells, as shown above, may be involved in regulation of these genes.
B1 methylation levels and gene expression. (A) Statistics for genomic occupancy of SINEs (squre), LINEs (circle), and LTR retrotransposons (triangle) in 1-kb bins are shown in relation to their distance from TSS for all (dashed lines) and testis-specific genes (solid lines). Testis-specific genes were listed previously (Weber et al. 2007). (B) Statistics for all and testis-specific genes that harbor 0 (white), 1 (gray), and >1 (black) B1 elements within 1 kb from TSS. (C) DNA methylation levels at B1 loci and expression levels of their neighboring genes. For each gene adjacent to a B1 locus, the relative proportion of mRNA levels in spermatogonia and liver (three biological replicates) was determined by quantitative RT-PCR.
To examine these possibilities, we compared the expression levels of genes possessing B1 elements in their TSS-proximal regions in spermatogonia and liver cells (Fig. 6C). Almost all genes examined showed elevated expression in spermatogonia over that in liver cells. In addition, the difference in expression was larger if the neighboring B1 was hypomethylated in spermatogonia but hypermethylated in liver cells (Pearson's r = 0.49 for differences in expression and neighboring B1 methylation). These results are consistent with a role for B1 elements in testis-specific gene expression.
Discussion
We examined the methylation levels at a panel of B1 loci in male germ cells at various developmental stages and in somatic tissue. In adult liver cells, most B1 loci were hypermethylated, consistent with the lack of B1 expression, although a small fraction located close to a Pol II promoter showed relatively lower methylation. Because human Alu elements show similar hypomethylation status around Pol II TSSs and in CpG-rich regions in somatic cells (Rodriguez et al. 2008; Xie et al. 2009; Edwards et al. 2010), this may be a common feature of mammalian SINEs. We found that this hypomethylation tendency of TSS-proximal B1 elements was much more evident in germ cells, especially during and after meiosis. Moreover, we showed that these B1-associated genes were highly expressed in germ cells, and, conversely, testis-specific TSS regions possess significantly more B1 elements than other TSS regions. These results suggest the possible involvement of B1 elements in regulation of testis-specific genes. B1 elements may play a role in stably silencing Pol II promoters in somatic cells, perhaps partly through methylation. Thus, a retrotranspositional insertion of a B1 element into a promoter region could modulate tissue-specific gene expression. Because B1 is retrotranspositionally active, this process may be ongoing in rodent evolution. In addition, the retrotransposed copies could then be another source of retrotransposition because of their tendency to be hypomethylated in germ cells.
We also showed that B1 loci in non-promoter regions were generally hypermethylated in spermatogenic cells. Our study with Pld6 and Piwil2 KO mice revealed that de novo methylation of B1 elements in prospermatogonia was largely independent of the piRNA pathway. This is in contrast to the piRNA-dependent de novo methylation of L1 elements (Aravin et al. 2007; Carmell et al. 2007; Kuramochi-Miyagawa et al. 2008, 2010; Watanabe et al. 2011a). Interestingly, methylation of the bulk of IAP elements was largely unaffected in Pld6 KO spermatogonia, although previous reports showed that two specific IAP loci were significantly affected in Pld6, Piwil2, and Piwil4 KO germ cells (Kuramochi-Miyagawa et al. 2008; Watanabe et al. 2011a). We also found that methylation of the bulk of MMERVK10C elements was unaffected, indicating that the involvement of the piRNA pathway in methylation was dependent on the retrotransposon class. Of note, the promoter of mouse L1 elements contains tandem repeats, whereas the promoters of IAP, MMERVK10C, and B1 elements do not. Therefore, it is conceivable that piRNAs (or processes generating piRNAs) play a more important role in targeted methylation of retrotransposon promoters consisting of tandem repeats of CpG-rich sequences. Consistent with this idea is the fact that the two IAP loci at which methylation depends highly on piRNAs have a specific segmental duplication of 80 bp (i.e., tandem repeat), which is absent from most IAP copies (data not shown). Moreover, the promoter of a non-coding RNA in the Rasgrf1 locus consists of CpG-rich tandem repeats, and its de novo methylation is highly dependent on piRNAs (Watanabe et al. 2011b), which also supports this idea.
In the mouse, de novo DNA methylation is catalyzed by DNMT3A and DNMT3B, and DNMT3L plays a role in assisting these two enzymes specifically in germ cells. Dnmt3l is indispensable for de novo methylation of the paternally methylated DMRs as well as the promoters of L1 and IAP retrotransposons (Bourc'his and Bestor 2004; Kaneda et al. 2004; Kato et al. 2007). Methylation of the H19 and Dlk1/Meg3 DMRs depends on the DNMT3A enzyme, whereas DNMT3A and DNMT3B have redundant functions in methylation of the Rasgrf1 DMR (Kaneda et al. 2004; Kato et al. 2007). To date, the mechanism underlying this discrepancy remains unknown.
Our analysis of B1 methylation revealed that methylation of the loci located in SINE-poor domains was more dependent on Dnmt3a, suggesting that its principal role is in methylation of SINE-poor genomic domains, which include H19 and Dlk1/Meg3, but not Rasgrf1. However, there remains a caveat. Methylation of the bulk of L1 elements enriched in SINE-poor regions did not show a significant Dnmt3a dependency. We speculate that the piRNAs assist DNMT3A and DNMT3B enzymes in methylating L1 elements regardless of their genomic locations.
SINEs are not distributed evenly along the chromosome arms. Rather, mammalian chromosomes consist of alternating SINE-rich/LINE-poor and LINE-rich/SINE-poor domains (Supplemental Fig. S6A), and SINE-poor genomic regions often coincide with chromosomal G bands, low G+C isochors, and gene-poor regions (Lander et al. 2001; Waterston et al. 2002). Among these genomic variables, we showed that the SINE density was most relevant to the observed variations in Dnmt3a dependency. We showed that SINE-poor/LINE-rich genomic regions were not spatially dispersed. Rather, they were localized at the periphery of prospermatogonial nuclei, as reported for somatic nuclei of humans and mice (Bolzer et al. 2005; Guelen et al. 2008; Solovei et al. 2009). Thus, an intriguing speculation is that DNMT3A can efficiently methylate the genomic DNA localized at the nuclear periphery, whereas DNMT3B cannot, although this is not the only possibility. In any event, our results shed light on the importance of discrete chromosomal domains for regulation of de novo methylation activities during male germ cell development.
Methods
Selection of B1 loci for methylation analysis
The RepeatMasker output and genome sequence were downloaded from the UCSC Genome Browser (http://www.genome.ucsc.edu/). We first selected B1 copies annotated as B1_Mm, B1_Mus1, or B1_Mus2, which were ≥135 bp, showed <9% divergence from the respective consensus sequences, and possessed ≥6 CpG sites. From this pool (consisting of ∼15,000 copies), we randomly selected loci (B1_001–B1_062). B1 loci in SINE-poor domains (0.5-Mb regions harboring <100 SINE elements; B1_101–B1_105) and promoter-proximal loci (at <1 kb from TSS; B1_201–B1_216) were randomly selected from the same pool. Their genomic locations are listed in Supplemental Table S1.
Cell preparation
Liver cells and spermatozoa were prepared separately from adult male C57BL/6J mice. Spermatogonia were isolated from P7 testes as follows: Seminiferous tubules were dissociated from testes by incubating in PBS containing 0.25% trypsin and 10 mM EDTA for 10 min at 37°C. The cell suspension was passed through a 40-μm nylon mesh and washed with PBS containing 10% fetal bovine serum (FBS). The cells were incubated with anti-EpCAM antibody (Santa Cruz Biotechnology) and Alexa Fluor 488 goat anti-rat IgG (Invitrogen), and the labeled cells were collected using FACS JSAN (Bay Bioscience). We confirmed that >95% of the sorted cells were positive for CD9 (BD Biosciences), another cell surface marker for spermatogonia. Pachytene spermatocytes and round spermatids were harvested from 5-wk-old testes. Seminiferous tubules were dissociated by incubating for 15 min at room temperature in PBS containing 50 U/mL of collagenase (Sigma), followed by incubation in PBS containing 0.05% trypsin for 20 min at 37°C. The cell suspension was then passed through a 40-μm nylon mesh and washed with PBS containing 10% FBS. After incubation in PBS containing 5 mM Vybrant DyeCycle Ruby stain (Invitrogen) and 10% FBS, the cells were sorted on the basis of their fluorescence emission intensity, which was correlated with the DNA content, to collect pachytene spermatocytes (4n) and round spermatids (1n). We confirmed high purity (>90%) for the pachytene spermatocyte preparation by immunostaining with anti-γH2A.X (Millipore) and anti-SCP3 (Abcam) antibodies. The round spermatid preparation was also almost homogeneous (>95%), as judged by immunostaining with anti-MVH antibody (Abcam).
DNA methylation analysis
Genomic DNA was prepared from cells using standard procedures (Sambrook and Russell 2001). For each cell type, DNA was isolated from three or four mice. Genomic DNA was subjected to bisulfite-mediated C-to-U conversion using BisulFast (Toyobo) and used as a template for PCR with ExTaq HS (Takara Bio). PCR comprised the following steps: (1) denaturation for 2 min at 95°C; (2) 20 cycles of touchdown PCR (a first cycle for 15 sec at 95°C, for 15 sec at 60°C, and for 30 sec at 72°C; during subsequent cycles, the annealing temperature was decreased by 0.5°C at each cycle); and (3) 15 cycles of PCR for 15 sec at 95°C, for 15 sec at 50°C, and for 30 sec at 72°C. The PCR products purified from an agarose gel were cloned into the pGEM-Teasy vector (Promega), and at least 12 clones (typically 15–20 clones) were sequenced for each B1 locus in each cell type. PCR primers and amplicon sequences for the B1 loci are listed in Supplemental Table S1. PCR primers for the analysis of autonomous retroelements are listed in Supplemental Table S2. Their methylation was analyzed using the respective consensus sequences in Repbase (http://www.girinst.org/repbase/) as a guide.
FISH analysis
LINE probes were prepared by PCR using green-dUTP (Abbott Molecular), unmodified dNTPs, and mouse genomic DNA as a template. For SINE probes, we prepared probes for the B1 and B2 sequences using Cy3-dUTP (GE Healthcare) and mixed equivalent amounts of the B1 and B2 probes. The primers for LINE (L1) PCR were described previously (Solovei et al. 2009). The primers for B1 PCR (fragment size of ∼108 bp) were 5′-GTGGTGGCGCACGCCT-3′ and 5′-TAGCCCTGGCTGTCCTGGAA-3′, and those for B2 PCR (fragment size of ∼151 bp) were 5′-GGCTGGTGAGATGGCTCAG-3′ and 5′-AGTACACTGTAGCTGTCTTCAGACAC-3′. Before use, the probes (10 ng/μL) were heat-denatured in 60% formamide, 2× SSC, and 50 mM sodium phosphate (pH 7.0). Dual FISH with SINE and LINE probes was performed on frozen sections as follows: The testes from E15.5 embryos were directly embedded in OCT compound and frozen on dry ice. The samples were then sectioned (6 μm) and fixed in 4% PFA. The complete hybridization protocol can be found at the following site: http://www.roche-applied-science.com/PROD_INF/MANUALS/InSitu/InSi_toc.htm. Germ cells were confirmed by immunostaining an adjacent section with anti-Oct-3/4 antibody (Santa Cruz Biotechnology).
RT-PCR analysis
Total RNA was isolated from the spermatogonia of P7 mice (FACS-sorted; three mice) and adult liver cells (three mice) using Isogene (Nippon Gene). cDNAs were synthesized from RNAs with random hexamers using a first-strand cDNA synthesis kit (Takara Bio) and used as a template for real-time PCR. The primer sequences are listed in Supplemental Table S3.
Acknowledgments
We thank Dr. Shin-ichi Tomizawa, Ms. Hatsune Chiba, Ms. Miho Miyake, and Mr. Yuichi Okudaira for their technical assistance. We also thank Drs. Yuji Kohara, Tomoko Tahira, and Hiroki Shibata for DNA sequencing. This work was supported by grants-in-aid to K.I. (22770009) and H.S. (20062010) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by The Naito Foundation to K.I.
Footnotes
-
↵9 Corresponding author.
E-mail ichiyanagi{at}bioreg.kyushu-u.ac.jp.
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.123679.111.
- Received March 23, 2011.
- Accepted August 23, 2011.
- Copyright © 2011 by Cold Spring Harbor Laboratory Press