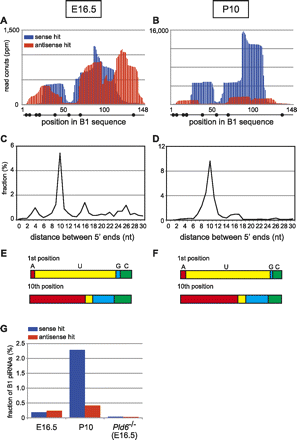
Analysis of B1-derived small RNAs in fetal and prepuberal testes. Small RNAs in E16.5 (Aravin et al. 2008) (ACE) and P10 (Aravin et al. 2007) (BDF) testes were mapped to B1_Mus1 consensus sequences. (A,B) Relative read counts (number of hits divided by the total sequence reads, expressed as parts per million, ppm) of sense (blue) and antisense (red) small RNAs. (C,D) The start position of each sense-strand read was compared with that of all antisense-strand reads. The percentages of relative length overlap between sense and antisense reads are shown. (E,F) Nucleotide compositions at the first and tenth positions of B1-derived small RNAs. (G) The relative abundance of B1-derived small RNAs. The fractions of sense (blue) and antisense (red) reads in the total sequence reads are shown. The data for piRNAs in Pld6/MitoPLD KO prospermatogonia were obtained from Watanabe et al. (2011a).











