Callipyge mutation affects gene expression in cis: A potential role for chromatin structure
- Susan K. Murphy1,2,
- Catherine M. Nolan3,
- Zhiqing Huang2,
- Katerina S. Kucera1,
- Brad A. Freking4,
- Timothy P.L. Smith4,
- Kreg A. Leymaster4,
- Jennifer R. Weidman1, and
- Randy L. Jirtle1,5
- 1 Department of Radiation Oncology, Duke University, Durham, North Carolina 27710, USA
- 2 Department of OB/GYN, Duke University, Durham, North Carolina 27708, USA
- 3 Zoology Department, University College Dublin, Belfield, Dublin 4, Ireland
- 4 U.S. Department of Agriculture, Agricultural Research Service, U.S. Meat Animal Research Center, Clay Center, Nebraska 68933-0166
Abstract
Muscular hypertrophy in callipyge sheep results from a single nucleotide substitution located in the genomic interval between the imprinted Delta, Drosophila, Homolog-like 1 (DLK1) and Maternally Expressed Gene 3 (MEG3). The mechanism linking the mutation to muscle hypertrophy is unclear but involves DLK1 overexpression. The mutation is contained within CLPG1 transcripts produced from this region. Herein we show that CLPG1 is expressed prenatally in the hypertrophy-responsive longissimus dorsi muscle by all four possible genotypes, but postnatal expression is restricted to sheep carrying the mutation. Surprisingly, the mutation results in nonimprinted monoallelic transcription of CLPG1 from only the mutated allele in adult sheep, whereas it is expressed biallelically during prenatal development. We further demonstrate that local CpG methylation is altered by the presence of the mutation in longissimus dorsi of postnatal sheep. For 10 CpG sites flanking the mutation, methylation is similar prenatally across genotypes, but doubles postnatally in normal sheep. This normal postnatal increase in methylation is significantly repressed in sheep carrying one copy of the mutation, and repressed even further in sheep with two mutant alleles. The attenuation in methylation status in the callipyge sheep correlates with the onset of the phenotype, continued CLPG1 transcription, and high-level expression of DLK1. In contrast, normal sheep exhibit hypermethylation of this locus after birth and CLPG1 silencing, which coincides with DLK1 transcriptional repression. These data are consistent with the notion that the callipyge mutation inhibits perinatal nucleation of regional chromatin condensation resulting in continued elevated transcription of prenatal DLK1 levels in adult callipyge sheep. We propose a model incorporating these results that can also account for the enigmatic normal phenotype of homozygous mutant sheep.
Callipyge (from the Greek: calli-, beautiful; pyge, buttocks) is the name of a muscle hypertrophy syndrome that affects some descendents of a ram (named Solid Gold) born to a flock of sheep in Oklahoma in 1983. This ram produced several offspring that exhibited enlarged muscles resulting from hypertrophy of fast-twitch muscle fibers, accompanied by a substantial decrease in total body fat. Investigation of the inheritance pattern of the callipyge phenotype revealed that it is only evident when transmitted from the sire; inheritance from an affected ewe does not result in callipyge. This suggested that genomic imprinting is somehow involved, since imprinted genes are expressed from only one allele in a parent-of-origin-dependent manner. Indeed, linkage mapping of the callipyge allele showed it was located at the distal end of ovine chromosome 18 (Cockett et al. 1994) in a region syntenic to an imprinted domain in humans and mice. However, callipyge is unique in that homozygous mutant sheep do not exhibit the muscle hypertrophy phenotype, indicating a more complicated biology than can be explained by standard imprinting models. This phenotype represents the first known example of what was then coined “polar overdominance” (Cockett et al. 1996).
We recently identified the mutation, a single A-to-G base change located intergenic to the two imprinted genes DLK1 (also referred to as PREF-1, FA1, pG2) and MEG3 (also referred to as GTL2) that gives rise to the callipyge phenotype (Fig. 1; Freking et al. 2002). The causality of this mutation was subsequently supported by an independent study (Smit et al. 2003). DLK1 is a paternally expressed gene encoding a member of the epidermal growth factor repeat-like family and is related to the delta-notch family of proteins (Laborda et al. 1993; Laborda 2000). MEG3 is expressed from the maternally inherited chromosome and encodes multiple alternatively spliced transcripts that do not contain obvious open reading frames and are therefore thought to function as noncoding RNAs (Miyoshi et al. 2000).
We and others have determined that genes contained within this domain (see Fig. 1), including DLK1, MEG3, DLK1 associated transcript (DAT), Paternally expressed gene 11 (PEG11; also referred to as Rtl1 in mice), and Maternally expressed gene 8 (MEG8), are imprinted in sheep (Charlier et al. 2001a,b; Murphy et al. 2005). While the presence of the callipyge mutation does not alter imprinting of these genes (Charlier et al. 2001a; Murphy et al. 2005), transcription levels of the genes linked in cis with the mutation are substantially influenced (Bidwell et al. 2001, 2004; Charlier et al. 2001a; Murphy et al. 2005), indicating a regional effect that is independent of imprinting.
Structure of the genomic region spanning the callipyge locus. Schematic representation of the telomeric end of ovine chromosome 18, and the genes present within the proximity of the callipyge mutation (A→G) on both the maternal (MAT) and paternal (PAT) chromosomes. Gene designations in mice are given in parentheses. The callipyge mutation (position 103,894 of AF354168) is contained within the transcribed CLPG1 locus. The direction of transcription is designated by the arrowheads representing each gene, with dark arrowheads representing actively transcribed alleles and translucent arrowheads representing silenced alleles for imprinted genes.
We previously reported detection of an RNA transcript, CLPG1 (Accession no. AF533009), that spans the region containing the callipyge mutation in longissimus dorsi muscle in sheep (Freking et al. 2002). Herein we investigate the expression of CLPG1 in fetal and adult sheep and the influence of the callipyge mutation on cytosine (CpG) methylation. We demonstrate that monoallelic expression of CLPG1 originates from the allele carrying the mutation in adult heterozygous sheep. Furthermore, the mutation hinders the acquisition of localized cytosine methylation in affected tissues of adult sheep in a manner inversely correlated with the number of mutated alleles present. Since CpG methylation is associated with condensed chromatin (Keshet et al. 1986; Antequera et al. 1989; Jones et al. 1998; Nan et al. 1998; Newell-Price et al. 2000; Eden et al. 2001; He et al. 2005), our data are consistent with the idea that the abnormal elevated expression of genes linked in cis with the mutation results from disruption of the normal perinatal formation of repressive chromatin structure in this domain.
Results and Discussion
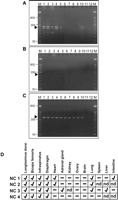
To begin to investigate how CLPG1 might be involved in the callipyge phenotype, we analyzed CLPG1 expression in tissues from four adult NMATCPAT (N and C refer to the normal and callipyge alleles, respectively, and the superscript designates the parental origin of the allele: MAT, maternally derived; PAT, paternally derived) sheep using RT-PCR (Fig. 2). We detected CLPG1 expression most consistently in muscles, including longissimus dorsi and biceps femoris, that are composed of fast-twitch fibers and undergo hypertrophy in the callipyge sheep. CLPG1 transcripts were also produced in infraspinatus, heart, and diaphragm muscles as well as in lung, adrenal gland, liver, and intestine. CLPG1 expression was not detected in kidney, ovary, brain, or spleen. The transcription of CLPG1 in infraspinatus of all four callipyge sheep indicates that CLPG1 RNA cannot be causal for the callipyge phenotype, since it is expressed in slow-twitch muscles that are unaffected in callipyge sheep.
Since imprinted genes flank CLPG1, we next determined whether CLPG1 is itself imprinted. We analyzed cDNAs from fetal and adult sheep heterozygous for the callipyge mutation (Fig. 3A; C, callipyge allele; N, normal allele; the maternal allele is listed first). In fetal sheep, CLPG1 was expressed from both parental alleles. When we analyzed the adult heterozygotes, we were surprised to find that CLPG1 is monoallelically expressed (Fig. 3B). Furthermore, CLPG1 is expressed from only the maternal allele in the CMATNPAT adults and conversely from only the paternal allele in the NMATCPAT sheep. We also analyzed expression of the transcript in biceps femoris, another hypertrophy responsive muscle, of two CMATNPAT and three NMATCPAT adults, and found that the transcript was likewise expressed monoallelically from the maternal and paternal alleles, respectively (data not shown).
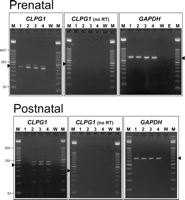
This unusual result prompted us to determine the expression status of this transcript in normal adult sheep. We identified several NMATNPAT sheep that were heterozygous for a G/A single nucleotide polymorphism (SNP) located 34 bp from the position of the callipyge mutation and also contained within the CLPG1 transcript. However, we were repeatedly unable to produce CLPG1 amplicons by RT-PCR from the longissimus dorsi muscle in these animals, while larger GAPDH amplicons were produced in each case (Fig. 4; data not shown). This was confirmed for a total of eight NMATNPAT animals and indicated that CLPG1 is normally not expressed in the NMATNPAT adults. When we examined CLPG1 expression in prenatal longissimus dorsi, we found that CLPG1 was, indeed, expressed in the NMATNPAT as well as the other three genotypes (Fig. 4). Although the homozygous CMATCPAT animals we examined were not informative at this second SNP, we did find that the CMATCPAT adults also express CLPG1 (Fig. 4). Together, these results suggest that CLPG1 is expressed during fetal development from both parental alleles, but expression is normally silenced postnatally in animals not carrying the callipyge mutation. Interestingly, the origin of nonimprinted monoallelic expression in the adult heterozygotes correlates with the presence of the mutation, suggesting that the mutation is somehow linked to CLPG1 transcription. This would predict that the CMATCPAT adult sheep express CLPG1 biallelically, but unfortunately because of the lack of polymorphisms in these inbred animals, it is not possible to confirm this at present.
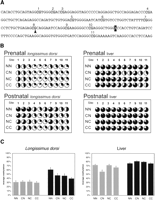
One established mechanism to regulate allele-specific gene expression is via cytosine methylation, with generally a positive correlation between gene silencing and methylation. Therefore, we next examined whether methylation differences were present across the CLPG1 region in all four genotypes that might account for the observed differential expression. The area encompassing the callipyge mutation is not classified as a CpG island using standard criteria (observed/expected > 0.6; average percent GC > 0.5; >200 bp in length). Nevertheless, it does contain several CpG dinucleotides that might contribute to the regulation of this region (Fig. 5A). We used bisulfite sequencing and PhosphorImager analysis to quantify methylation of the CpG dinucleotides in the immediate vicinity of the mutation in two prenatal and two postnatal animals of each genotype (Fig. 5B,C). Figure 5B shows the average methylation detected for each CpG position, while the bar graphs in Figure 5C show the average methylation detected for all CpGs combined. During fetal development, there were no significant differences between the genotypes in longissimus dorsi when the average percent methylation for the entire region was calculated (ANOVA P = 0.9289). However, in normal adult sheep (NMATNPAT), there was an approximate twofold increase in the level of methylation compared to that seen in the fetal NMATNPAT sheep (63.2% vs. 32.1%, respectively), indicating perior postnatal acquisition of increased methylation for these CpG dinucleotides. Remarkably, we did not observe the same proportionate increases in methylation in the adult sheep carrying the callipyge mutation (Fig. 5B,C). Compared to the NMATNPAT adults, the heterozygotes exhibited intermediate levels of methylation (CMATNPAT, 48.48%, P = 0.05; NMATCPAT, 46.59%, P = 0.04), and the homozygous adult CMATCPAT sheep have the lowest methylation level (39.88%, P = 0.001 vs. NMATNPAT adults; ANOVA P = 0.01). These striking results indicate a postnatal hindrance in the ability to acquire de novo methylation across this genomic locus in the presence of the mutation.
Expression of CLPG1 in tissues of an adult callipyge sheep. (A) RT-PCR products generated using a gene-specific primer for CLPG1 in the reverse-transcription step. CLPG1-specific amplicons are 270 bp and are designated by the arrowhead. The other bands are nonspecific products of the amplification, confirmed by sequencing. (B) Controls for each reaction in which the reverse transcriptase was omitted. (C) GAPDH amplicons (452 bp) produced using oligo(dT)-primed cDNA from the same RNA samples as those used in A and B. (A,B,C) (lane M) 50-bp marker; (lane 1) longissimus dorsi; (lane 2) biceps femoris; (lane 3) infraspinatus; (lane 4) diaphragm; (lane 5) heart; (lane 6) adrenal gland; (lane 7) kidney; (lane 8) ovary; (lane 9) brain; (lane 10) lung; (lane 11) empty lane; (lane 12) water control. (D) Detection of CLPG1 transcripts in four adult callipyge sheep (NC 2 results represent those shown in panel A). A check-mark indicates detection of the CLPG1 transcript in the designated tissue; a minus sign indicates the transcript was not detected; (nd) not determined.
Imprint analysis of CLPG1 in longissimus dorsi. (A) Nucleotide sequence (using a reverse sequencing primer) of cDNAs generated from two CMATNPAT (CN) and two NMATCPAT (NC) fetal sheep showing expression of the CLPG1 transcript from both the maternal and paternal alleles, evidenced by the presence of both the T (normal) and C (callipyge) bands at the position of the callipyge mutation (arrowheads). (B) Nucleotide sequence of gDNA and corresponding cDNAs for three CMATNPAT and three NMATCPAT adult sheep. The gDNA shows the presence of both the T (normal) and C (callipyge) alleles (arrowheads), while the cDNA sequence shows only that the C allele is expressed in adult sheep, regardless of the parental inheritance for that allele.
Prenatal and postnatal expression of CLPG1 in sheep of all four genotypes. RNA from each individual was reverse-transcribed in the presence or absence (no RT) of reverse transcriptase using a gene-specific primer, followed by PCR amplification and resolution by agarose gel electrophoresis. The expected amplicons are 270 bp and are designated by the arrowheads. GAPDH amplicons (452 bp) were amplified from the same tissues. (Lane M) 50-bp marker; (lane 1) NMATNPAT; (lane 2) CMATNPAT; (lane 3) NMATCPAT; (lane 4) CMATCPAT; (lane W) water control; (lane E) empty lane.
Because callipyge is specific to fast-twitch muscles, we hypothesized that the observed methylation patterns would only be observed in affected tissues if they have relevance to callipyge. We therefore examined the methylation status of these same sites in liver tissues from prenatal and postnatal animals of all four genotypes (Fig. 5B,C). The methylation level is much higher overall in liver than in longissimus dorsi, with normal prenatal sheep exhibiting an average 72.5% methylation, while postnatal sheep exhibit an average 76.3% methylation. Unlike longissimus dorsi, there is not a perinatal doubling of the methylation levels across this locus in liver, and there are not significant differences in methylation between the genotypes of adult sheep (ANOVA P = 0.19). These results indicate that the attenuation of methylation acquisition observed in longissimus dorsi for this locus is specific to the affected tissues and results from the presence of the mutation.
One confounding aspect of the underlying mechanism of callipyge is the apparently normal phenotype of the homozygous CMATCPAT sheep. Georges et al. (2003) proposed that trans interactions between the genes or gene products within the callipyge region would account for the incomplete penetrance of the phenotype in these animals. Our results support this contention. As shown schematically in Figure 6, we believe that MEG3 may act in trans to down-regulate DLK1 expression, since we previously determined that the expression of DLK1 relative to MEG3 is elevated in the affected adult animals and is highly correlated with the callipyge phenotype (Murphy et al. 2005). This is also supported by the finding that mice inheriting a deletion of the imprint control region upstream of Gtl2 (MEG3) on the maternal allele exhibit loss of imprinting for all examined genes in the domain. In this case, Gtl2 expression is abrogated, as are other maternal noncoding RNAs in the domain, while Dlk1 expression is up-regulated and originates from both parental alleles, suggesting that transcription of Gtl2 or other maternally expressed genes is required to regulate expression of Dlk1 (Lin et al. 2003). Others have also postulated a role for PEG11 (Rtl1) and RNA interference originating from transcripts of the PEG11-AS (Rtl1-AS) locus in the callipyge phenotype (Bidwell et al. 2004; Davis et al. 2005), although increasing evidence points toward DLK1 as the effector of callipyge (Davis et al. 2004; Kim et al. 2004; Murphy et al. 2005).
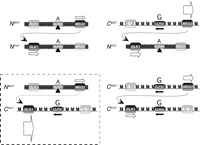
Based on our results here, we refine this model by incorporation of the concept that chromatin structure, altered by the presence of the callipyge mutation, is integrally involved in mediating the callipyge phenotype. We have shown that in the NMATNPAT adult sheep, the intergenic DNA at the CLPG1 locus exhibits elevated postnatal cytosine methylation, presumably reflecting the results of chromatin remodeling. This is accompanied by CLPG1 silencing and a reduction in DLK1 mRNA levels (Murphy et al. 2005). In the NMATCPAT animals, the presence of the mutation on the paternally inherited chromosome prevents effective chromatin remodeling, and we have shown that CLPG1 and DLK1 are expressed at abnormally elevated levels in adulthood. In this case, MEG3 transcripts produced from the nonmutated maternally inherited chromosome are insufficient to abrogate DLK1 expression in trans, resulting in the callipyge phenotype. For the CMATCPAT sheep, expression of MEG3 is enhanced, as would also be expected for DLK1 because of the homozygous mutational status. In these animals, however, the elevated levels of MEG3 transcripts control DLK1 expression in trans, and thus these sheep do not have the callipyge phenotype. Our results further suggest that this regulation occurs at the transcriptional level, since the abundance of DLK1 mRNAs in the CMATCPAT sheep is equivalent to that observed in NMATNPAT sheep (Murphy et al. 2005).
The monoallelic transcription of CLPG1 only from the mutated allele in the adults suggests that the mutation is somehow allowing for constitutive regional gene expression from the affected allele. This is supported by the observation that while both DLK1 and MEG3 remain imprinted, transcription in adult animals is substantially increased from the allele carrying the mutation (Murphy et al. 2005). This long-range effect is consistent with the notion that the mutation may affect enhancer/silencer function, as previously proposed (Georges et al. 2003). Alternatively, it may affect chromatin structure with regional consequences, as we hypothesize here (Fig. 6). Increased postnatal methylation concomitant with decreased DLK1 and CLPG1 transcription in normal adult sheep suggests that chromatin structure is an important regulatory mechanism in the control of gene expression for this domain. Our results indicate that the callipyge mutation causes postnatal hypomethylation of this locus and prohibits down-regulation of CLPG1 transcription. This single base change also causes enhanced transcription of genes linked in cis to the mutation but does not cause defects in imprinting. We therefore hypothesize that the callipyge mutation modifies binding of a fast-twitch muscle-specific protein or protein complex that serves to nucleate regional chromatin condensation. As a result, elevated transcription is observed for genes within reach of this effect including DLK1, MEG3, and CLPG1. In support of this hypothesis, Charlier (2004) recently reported detection of a protein in sheep that binds to the region in which the callipyge mutation is located, and that binding of this protein is reduced when the mutation is present. Further work will be required to determine the identity of this protein and its role in the callipyge phenotype.
Methylation analysis of the genomic region flanking the callipyge mutation. (A) Nucleotide sequence encompassing the position of the callipyge mutation (highlighted A) showing the CpG dinucleotides analyzed. (B) Quantitative CpG methylation analysis in prenatal and postnatal longissimus dorsi and liver tissues from sheep of each genotype. The status of each CpG is shown by the individual pie charts, where the darkened portion corresponds to the average level of cytosine methylation detected for N = 2 animals of each genotype. CpG position 8 within the sequence in A was omitted from analysis because of the presence of a G→A SNP (arrowhead in A) in some animals. (NN) NMATNPAT; (CN) CMATNPAT; (NC) NMATCPAT; (CC) CMATCPAT. (C) Locus-specific methylation for all CpGs (except position 8) within the sequenced region in longissimus dorsi and liver of prenatal and postnatal sheep. Shown is the average percent methylation (±SEM) across the 10 CpGs within the amplicon for two sheep of each genotype. (Gray bars) Fetal sheep; (black bars) adult sheep.
At this point, it would appear that the CLPG1 transcript itself does not have a causal role in the callipyge phenotype, since it is expressed in unaffected infraspinatus of the callipyge adult sheep. It is likely that the gene products produced from this locus are not functionally significant but rather are simply the result of open chromatin and the associated permissive conditions for transcription. Indeed, we have also detected an as-yet-uncharacterized transcript produced across this same region but from the opposite coding strand using gene-specific primers for reverse transcription (data not shown). Our data at this point are consistent with the mutation causing regional deregulation of gene expression through induced chromatin alterations.
These data have helped illuminate some of the enigmatic aspects of the callipyge phenotype and have contributed to our understanding of how a seemingly innocuous SNP has the potential to induce a considerable phenotypic change. Future work will be required to discern whether and how MEG3 (or other gene products within the callipyge domain) regulates DLK1 in trans and the specific nature of the hypothesized chromatin alterations induced by the presence of the mutation. Whether callipyge can be modeled (e.g., in mice engineered to carry the mutation) to further address questions about the biological basis for the effects of the callipyge mutation remains to be determined.
Methods
Tissues
Fetal and adult sheep tissues from all four callipyge genotypes were obtained from flocks maintained at the United States Department of Agriculture Meat Animal Research Center in Clay Center, Nebraska. Matt Doumit (Department of Animal Science, Michigan State University, East Lansing, MI) kindly provided additional muscle tissues from adult CMATNPAT animals from the same flocks, for confirmation of the allelic expression of CLPG1.
Nucleic acid purification
Genomic DNA was prepared from frozen tissues by proteinase K digestion (Qiagen) followed by standard phenol:chloroform extraction with Phase Lock Heavy gel separation tubes (Eppendorf) and ethanol precipitation. Total RNA was isolated from frozen tissues using RNA Stat-60 according to the manufacturer's directions (TelTest).
Methylation analysis
Genomic DNA was modified by conversion of unmethylated cytosines to uracils using sodium bisulfite as previously described (Grunau et al. 2001; Murphy et al. 2003). Bisulfite-treated DNA was subjected to PCR amplification using primers BS-F1 (5′-GGTGATTAGTAGTAGAGTAGAGATA-3′) and BS-R1 (5′-CTATTTAATCCTAAATAAATCCTCT-3′) (94°C for 3 min, 40 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec, then 5 min at 72°C). The amplicons were resolved on agarose gels and purified using Sigma GenElute spin columns (Sigma Aldrich).
Mechanistic model for the callipyge mutation. Schematic representation of the four genotypes at the callipyge locus in adult sheep, the patterns of gene expression for DLK1, MEG3, and CLPG1, and a model for chromatin structure of each allele based on our current and prior findings (Murphy et al. 2005). The relative position of the mutation within the CLPG1 locus is shown above each allele (A, normal; G, mutated). Arrows beneath transcribed DLK1 and MEG3 are proportional to transcription levels (Murphy et al. 2005). In adult sheep, the ratio of DLK1:MEG3 transcripts is highly correlated with phenotype in a temporal and tissue-specific manner, suggesting that increased MEG3 transcription is necessary for the postnatal down-regulation of DLK1. In normal sheep, attenuation of DLK1 transcription is accompanied by a doubling of cytosine methylation in the region of the callipyge mutation. Reciprocal heterozygotes and CMATCPAT exhibit deficient cytosine methylation, and this may reflect a more open chromatin structure and permissiveness for transcription. This is supported by the observed monoallelic expression of CLPG1 only from alleles carrying the mutation (black arrows). The mutation may therefore alter the ability of regional chromatin to condense perinatally via inhibition of chromatin remodeling complex assembly (black triangles), inhibition of DNA methylation, and/or altered histone modifications.
To determine the overall level of methylation, nucleotide sequence analysis was performed using the Radiolabeled Terminator Cycle Sequencing Kit (USB Corporation) with primer BS-F2 (5′-GGTTTGTTTATATTTGTAGTAGG-3′), followed by polyacrylamide gel electrophoresis and determination of the percent cytosine methylation—(methylated C)/[(unmethylated C) + (methylated C)]—by PhosphorImager analysis (Molecular Dynamics Storm with ImageQuant software; Amersham Biosciences). A G/A polymorphism is present at one of the CpG dinucleotides within the region amplified by the primers (position 103,860 of Accession no. AF354168), and the recent generation of the callipyge allele occurred on a haplotype containing the A allele from this nearby single nucleotide polymorphism. The presence of the A at this position prevents methylation of the adjacent cytosine, and therefore analysis of the methylation status of this dinucleotide was not included in determining the methylation level of this region (Fig. 5C). Statistical comparisons across groups were performed using One-way Analysis of Variance (ANOVA) and the Dunnett Multiple Comparisons Tests (InStat; GraphPad Software). Comparisons between genotypes were performed using unpaired Student's t-tests with a two-tailed distribution. P-values <0.05 were considered significant.
CLPG1 expression analysis
Using the callipyge mutation (position 103,894 of AF354168) and an upstream SNP as markers (position 103,860 of AF354168), heterozygous sheep were analyzed for allelic expression of CLPG1 by comparisons of the nucleotide sequence of genomic DNA to matched cDNA. Total RNA was treated with DNase I (Invitrogen) and subjected to reverse transcription in the presence and absence of reverse transcriptase (Superscript II; Invitrogen) using a strand-specific primer (5′-CCAGGAGGCTGC CAGGAGACC-3′), followed by PCR using forward primer F2 (5′-CAGATGCT GTGGACGTGGGGAATC-3′) and reverse primer R244 (5′-GCAAGGGTCT GTTTGGTCCTAA-3′) with conditions as follows: 94°C for 3 min; then 45 cycles of 94°C for 30 sec, 66°C for 30 sec, 72°C for 30 sec; and a 5-min extension at 72°C. GAPDH was used as an internal control and was amplified from oligo(dT)-primed cDNA generated from the same RNA preparations as those used for the CLPG1 analysis. Forward primer GAPDH-452F (5′-ACCACAGTC CATGCCATCAC-3′) and reverse primer GAPDH-452R (5′-TCCACCACCCTGTT GCTGTA-3′) were used to generate 452-bp amplicons from cDNA as follows: 94°C for 3 min; then 35 cycles of 94°C for 30 sec, 60°C for 30 sec, 72°C for 30 sec; then a 5-min extension at 72°C. The 270-bp CLPG1 products were resolved by agarose gel electrophoresis, purified using Sigma GenElute columns (Sigma-Aldrich), then sequenced using the Radiolabeled Terminator Cycle Sequencing Kit (USB Corporation) with primer R3 (5′-GAT CACCCACACCCTGCC-3′). Sequencing products were resolved on 5% polyacrylamide sequencing gels and exposed to radiographic film and/or to a PhosphorImager screen for analysis using ImageQuant software (Amersham Biosciences).
Acknowledgments
We thank Matt Doumit from the Department of Animal Science, Michigan State University, East Lansing, Michigan for kindly providing additional CMATNPAT adult sheep tissues for confirmation of the expression status of CLPG1 transcripts. We also thank Ali Gusberg at Duke University (Durham, NC), and Renee Godtel and Kevin Tennill at the MARC location (Clay Center, NE) for excellent technical support contributions. This research was supported by NIH grants CA94668 to S.K.M., Enterprise Ireland grant IC/2003/20 to C.M.N., and CA25951, ES08823, and ES13053 to R.L.J.
Footnotes
-
Mention of a trade name, proprietary product, or specified equipment does not constitute a guarantee or warranty by the USDA and does not imply approval to the exclusion of other products that may be suitable.
-
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4389306.
-
↵5 Corresponding author. E-mail jirtle{at}radonc.duke.edu; fax (919) 684-5584.
-
- Accepted November 21, 2005.
- Received July 6, 2005.
- Cold Spring Harbor Laboratory Press