Identification of Wound Healing/Regeneration Quantitative Trait Loci (QTL) at Multiple Time Points that Explain Seventy Percent of Variance in (MRL/MpJ and SJL/J) Mice F2 Population
Abstract
Studies on genetic mechanisms of wound healing in mammals are very few, although injury is a leading cause of the global burden of disease. In this study, we performed a high-density, genome-wide scan using 633 (MRL/MPJ × SJL/J) F2 intercross at multiple time points (days 15, 21, and 25) to identify quantitative trait loci (QTL) involved in wound healing/regeneration. The hypothesis of the study was that QTL and unique epistatic interactions are involved at each time point to promote wound healing/regeneration. Ten QTL were identified from chromosomes 1, 4, 6, 7, 9, and 13. Of the 10 QTL, eight from chromosomes 1, 4, 6, and 9 were novel as compared to QTL identified in the McBrearty et al. (1998) study. The 10 QTL altogether explained 70% of variance in F2 mice. The same QTL were identified at each time point, with simple linear correlation between days 15, 21, and 25, showing very high significant relationships (R >0.92,P <0.0001). Unique epistatic interactions were identified at each time point except those from chromosomes 4, 6, 9, and 13 that were found at all three time points, showing that some loci are involved at all the three time points of wound healing (days 15, 21, and 25). Therefore, loci-to-loci interactions may play a major role in wound healing. Information from these studies may help in the identification of genes that could be involved in wound healing/regeneration.
Injury is a leading cause of the global burden of disease that occurs in the battlefield for army personnel and as accidents in civilian populations. It is estimated to cost healthcare providers in excess of $500 billion per year in the United States alone to treat injuries (Satcher 2000). Therefore, there is an urgent need to develop effective strategies to prevent injuries and promote wound healing after injuries. However, poor understanding of molecular mechanisms underlying fast wound healing/regeneration process has impeded therapeutic treatment of tissue injury.
The primary goal of wound treatment is rapid wound closure with minimal scar formation, which is important for cosmetic appearance (Goss 1992;Martin 1997; Singer and Clark 1999). Wound healing has been accelerated by use of certain proteins like cod liver oil ointment, vitamin A, and deoxyribonucleosides (Shan et al. 1995; Chen et al. 1999; Terkelsen et al. 2000). Animal models have been developed to study mechanisms of wound healing in normal or impaired states (Rothe and Falanga 1992). Although molecular and cell biology studies have explained some of the biological processes that could be involved in wound repair (Rothe and Falanga 1992; Singer et al. 1999), we still know very little about wound healing, as it is a complex process involving many genes (Rothe and Falanga 1992; Heber-Katz 1999; Kunimoto 1999; Trengove et al. 2000).
A recent study in our laboratory (Li et al. 2001b) and an earlier study by Clark et al. (1998) indicated that the MRL/MpJ-Faslpr(MRL-F) strain of mice could completely heal an ear-punched hole (2 mm in diameter) within 30 d with no scar tissue. In contrast, the C57BL/6 strain of mice healed only 40% and SJL/J <25% of the original hole with scar tissue at the same length of time. To date, McBrearty et al. (1998) have demonstrated that rapid wound healing in MRL-F mice is a genetically controlled quantitative trait. The McBrearty et al. (1998)study used MRL-F and C57BL/6, F2 intercross at one time point, which resulted in the identification of five quantitative trait loci (QTL) explaining unknown percentage of variance in F2 mice.
This pioneer study (McBrearty et al. 1998) has demonstrated that MRL mouse is an ideal model to elucidate the molecular mechanisms that underlie wound-repair/regeneration in mammals. To further identify genetic mechanisms controlling wound healing, we used F2population from progenitor strains that have different genetic origin (MRL/MpJ and SJL/J). The objectives of this study were to map fast-healer genes using two genetically extreme progenitor strains (MRL/MpJ and SJL/J) and to identify epistatic interactions at multiple time points. In this report, we present data showing the identification of same QTLs at each time point that explain 70% of the variance in F2 mice and that QTL together with epistatic interactions could promote wound healing.
RESULTS
Wound Healing in Progenitor Mice (MRL/MPJ and SJL/J) From Day 1 to Day 40 and the Distribution of Hole Size in F2 Mice Population at Day 21
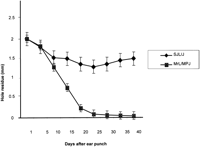
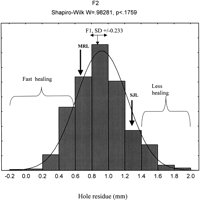
To study the genetic mechanisms of wound healing, two progenitor strains (MRL/MpJ and SJL/J) with extreme phenotype were needed for F2 intercross. Figure 1 shows the comparison of wound healing in MRL/MpJ (fast healer) and SJL/J (slow healer) progenitor strains. MRL/MpJ heals fast and closes the 2-mm hole within 30 days while SJL/J heals <25% at the same period of time, indicating that MRL and SJL are good models to study molecular mechanisms of wound healing. Figure 2 shows the distribution of wound healing size at day 21, demonstrating a normal distribution indicating that the ear phenotype is multigene controlled. The distribution also demonstrates that some F2mice heal faster than MRL/MPJ progenitor (fast healer) and others heal less than SJL/J (slow healer) (shown by arrows). The results show large differences in wound healing between MRL and SJL progenitor mice, and phenotype-dependent F2 distributions demonstrate the inherited nature of the rate of wound healing. As shown in Figure 2, the F1 have a mean hole size (0.8 mm) that is intermediate of the progenitor strains with a standard deviation of 0.233. Therefore, these data are ideal for use in quantitative-trait loci studies to map genes for wound healing.
Rates of healing of 2-mm ear hole in MRL (fast healer) and SJL (slow healer) progenitor mice at different timepoints (days 1–40). TheY-axis shows the hole residue in millimeters (mm), and theX-axis shows the number of days after the ear punch.
The histogram of frequencies of the hole residue size on day 21 after wounding in F2 (MRL/MpJ × SJL) mice population. Arrows indicate F2 that heal better than MRL/MpJ and F2that heal worse than SJL/J progenitor mice. The Y-axis shows the number of observations, and the X-axis shows the hole residue in millimeters (mm).
Use of High-Density Genome-Wide Scan to Identify QTL at Different Timepoints in 633 (MRL × SJL) F2 Mice
To increase the power of the study to detect QTL, we used 119 markers and 633 F2 mice. Over 560 microsatellite markers were screened for polymorphisms between MRL/MpJ and SJL/J mice. One hundred and fifty eight markers were informative between the two progenitor strains of mice. After the first round of QTL loci screen, extra markers were added within the QTL regions to increase the accuracy of QTL position. The markers then were chosen depending on their position on the chromosome in an effort to distribute them at about 15 cM or less to generate a complete genome-wide scan. The 119 markers were used to detect QTL involved in wound healing/regeneration in 633 (MRL/MpJ × SJL/J) F2 mice intercross at multiple time points. The order of these markers was as predicted by the available genomic maps (Whitehead Institute, Massachusetts Institute of Technology and Jackson Laboratory, http://www.jax.org).
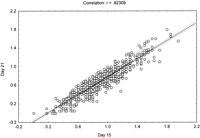
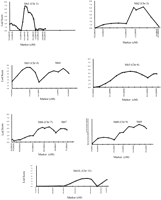
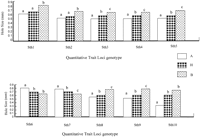
Multiple timepoints were examined in the present study to determine if different QTL are involved at each stage of wound healing. Table1 shows that the same QTL were identified at days 15, 21, and 25. By simple linear correlation, significant relationships were noted between hole residue at days 15 and 21 as shown in Figure 4(R = .92; P <.0001). Similar regression analyses (data not shown) were performed at days 21 and 25 (R = .97;P <.0001) and days 15 and 25 (R = .90; P<.0001), and very strong intraclass correlation of 0.74, indicating that some of the QTL could be involved in wound healing at all three timepoints. Table 1 and Figure 3 show a total of nine significant QTL (logarithm of the odds [LOD] score ≥3.5) and a suggestive QTL on chromosome 13 (LOD score ≥2.7). Figure 3 shows histograms of 10 QTL at day 21, which are the same as those at days 15 and 25. The QTL were identified on chromosomes 1, 3, 4, 6, 7, 9, and 13. Chromosomes 4, 7, and 9 had two QTL each as shown in Figure 3 and Table 1. The 10 QTL explain 70% of variance in F2 mice as shown in Table 1. The 10 QTL are distributed along seven chromosomes with soft tissue heal 3 (Sth3) and Sth4 on chromosome 4, Sth6 and Sth7 on chromosome 7, and Sth8 and Sth9 on chromosome 9. Because one quantitative trait locus is fitted to the model at a time, the estimated effects of the two QTL on the same chromosome may be biased. Figure 5shows peak loci that exhibited significant/suggestive associations between the genotypic markers and phenotypic values. Out of the 10 QTL, eight MRL homozygote genotypes contributed more in promoting wound healing than SJL homozygote (except for Sth6 QTL in which SJL homozygote contributed more to promote wound healing than MRL homozygote, Fig. 5, bottom). The alleles at four out of the 10 QTL showed a dominant inheritance (Sth1, Sth2, Sth6, and Sth10) while the remaining six QTL demonstrated an additive inheritance. Wound healing may be influenced not only by these QTL but also by epistatic interactions between different loci in the QTL.
Location of Sth QTL as Determined from the F2 Intercross
Ear hole size (mm) at day 15 was regressed against ear hole size (mm) at day 21. The correlation coefficient and the significance level are indicated. All the other comparisons (day 15 against day 25 and day 21 against day 25) showed the same high significance level.
Interval mapping at day 21, showing significant quantitative trait loci (QTL) (1,3,4,6,7, and 9) and a suggestive QTL on chromosome 13. The logarithm of the odds (LOD) score is on the Y-axis and relative location of marker name and distribution in centiMorgans (cM) on the X-axis. The QTL are labeled as soft tissue heal (Sth) at each peak.
The effects of genotype on hole size in mm. The genotypes were derived from the markers near the quantitative trait loci (QTL) peaks. The capital letters A and B represent homozygote genotype for Mrl/MPJ and SJL/J mice, respectively, and H represents heterozygotes genotype. The lower case letters on the top of bars indicate the level of difference in phenotypic value between genotypes. If the two bars share the same letter, the value between the two genotypes will not be statistically significant, but if different, then they are statistically different atP<.05.
Epistatic Interactions Within Different QTL Promote Wound Healing in (MRL × SJL) F2 Mice
Epistatic interactions could be involved in wound healing at each time point (days 15, 21, and 25). Table 2shows 17 different epistatic interactions between different loci. A two-way ANOVA model was used to identify interactions, which were significant at P <0.05. Five loci had epistatic interactions at day 15 only, 2 loci at day 15 and 21, three loci at days 15, 21, and 25, while three loci were significant at days 21 and 25, and lastly, four loci were only significant at day 25. All the significant loci at day 21 also were found either at day 15 or day 25. Day 21 is a transition between fast wound healing and remodeling stages, therefore an overlap of loci interaction is expected. There were unique epistatic interactions at each time point, though some interactions were present at days 15, 21, and 25, indicating that some loci are involved at all time points of wound healing tested.
Loci that Produce Genetically Significant Interactions by a Two-Way ANOVA Analysis
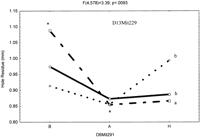
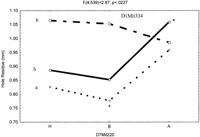
To determine if allelic variation contributes to wound healing at each of the significant loci, a two-sided t-test was performed on the three allelic (A, B, H) combinations. The data presented in Figure6 show two examples of epistatic interactions to illustrate the different effects of allelic combination. Figure 6A shows an epistatic interaction between loci on Chrs 6 and 13, represented by markers D6mit291 and D13mit229. It is described as an interaction between the additive effects from Chrs 6 and Chrs 13, leading to better wound healing. Overall, out of the eight possible allelic combinations, allele AA combination significantly promotes wound healing (P <.03). Mice with ‘Aa’ allele combination healed 57% of the wound while those carrying ‘Ba’ allele only 46% (P <.0005, indicated by *). Secondly, Figure 6B shows loci interaction between D1Mit334 and D7Mit220 on Chrs 1 and 7. Mice carrying ‘Aa’ allelic combination generally do not significantly contribute to better wound as compared to the mice with other eight allelic combinations. F2 mice with allele combination of ‘Ba’ heal 62% of the wound as compared to mice with ‘Ah’ allele combination, which heal only 47% (P <.008, indicated by *).
The interactive effects of the QTLs, with three alleles, A (MRL), B (SJL) and H (heterozygotes). (A) Shows epistatic interaction between loci D6Mit291 and D13Mit229. The Y-axis (leftside) shows hole residue in millimeters (mm), Y-axisright side loci D13Mit229 (a, h, and b) and theX-axis shows loci D6Mit291 (B, H, and A). The * means significantly different from the two extreme allelic combinations (Aa and Ba). (B) Shows epistatic interaction between loci D1Mit334 and D7Mit220. The Y-axis (left side) shows hole residue in millimeters (mm), Y-axis right side loci D1Mit334 (a, h, and b) and the X-axis shows loci D7Mit220 (H, B, and A). The * means significantly different from the two extreme allelic combinations (Ba and Ah alleles).
DISCUSSION
Wound healing occurs in three stages, inflammatory (0–2 d), fast wound healing (5–20 d), and remodeling stages (>21 d) (Martin 1997;Singer et al. 1999). In this study, we examined days 15, 21, and 25 with the hypothesis that genes involved in wound healing could be different at each time point. The major findings of these studies were (1) 10 QTL were identified, eight novel and two previously identified by McBrearty et al. (1998), together explain 70% of the variance in wound healing of F2 MRL/SJL intercross, and surprisingly, all the QTL were identical at each time point, and (2) some epistatic interactions were unique for each wound-healing stage, while others were common at all time points. These findings will be discussed sequentially.
The Same QTL Were Identified at Each Timepoint and Together Explained 70% of Variance in Wound Healing in the (MRL/SJL) F2 Mice
To identify QTL that may carry genes that are involved in fast wound healing, MRL-MPJ and SJL/J mice were selected, as they have extreme ear healing capacity. The phenotype shows a normal distribution in F2 mice intercross (MRL-MPJ × SJL/J), a distribution that indicates that the genes regulating fast wound healing have additive effects and no major gene with dominant inheritance exists in the segregating population, features consistent with polygenic inheritance.
We used (MRL × SJL/J) F2 cross to identify 10 QTL, eight were novel and two were identified previously by McBrearty et al. (1998). Because of the different genetic origin between progenitor strains (MRL and SJL/J), our study identified eight QTL that could not be identified using MRL × C57BL/6 combination (McBrearty et al. 1998). Our study used the same fast healer, MRL, as in McBrearty et al. (1998), but a different slow healer, SJL/J. The QTL on chromosome 7 and a suggestive QTL on chromosome 13 were identified also by McBrearty et al. (1998) using MRL × C57BL/6 strain combination. To our surprise, one of the two QTL on chromosome 7 (D7Mit220) is contributed by SJL/J, a poor healer, indicating that even poor healers that do not express the trait are important in the promotion of wound healing (Frankel 1995; McBrearty et al. 1998). These findings show that the major contributor to wound healing is MRL, as it is the common strain between this study and the McBrearty et al. (1998) study. The 10 QTL explained 70% of variance in F 2 mice. As explained in the results section on the percentage explained by variance, the QTL analysis program fitted only a single model each time so there could have been an overestimate on the chromosomes that had more than one quantitative trait locus. A very conservative value may be between 50% and 70%. To our knowledge, there is no other mouse QTL study that has explained such a high percentage of variance for any phenotype. The reasons could be partly because of high marker density genome-wide scan, which increased the QTL detection power (Moore and Nagle 2000).
We hypothesized that different QTL would be identified at different time points as a result of different genes that may be involved at each stage of wound healing/regeneration (Martin 1997; Singer et al. 1999). Surprisingly, the same QTL were identified at each time point, with very high phenotypic correlation between days 15, 21, and 25, and intraclass correlation, suggesting that some genes could be involved in the three time points tested. There is evidence that some genes are consistently up-regulated at all stages of wound healing, most of which are growth factors and have well-characterized roles in wound healing (Iyer et al. 1999; Li et al. 2001a,c). An example of these genes are insulin-like growth factor 1(IGF-1), insulin-like growth factor binding protein 1 (IGFBP-1), IGF-1 receptor, transforming growth factor β 2 (TGFβ2), fibroblast growth factor 7 (FGF 7), fibroblast growth factor receptor 2 (FGFR 2), vascular endothelial growth factor (VEGF), and bone morphogenetic protein 4(BMP4) (Li et al. 2001c). This indicates that growth factors are involved in all stages of wound healing, which solidifies our suggestion that some genes participate at the three timepoints of wound healing.
Epistatic Interactions Could be Important in the Promotion of Wound Healing in (MRL-MPJ x SJL) F2 Mice Intercross
In this study, we hypothesized that lower LOD scores (<2.7) will show significant interactions. A two-way ANOVA test model was used in genome wide scan to identify 17 epistatic interactions. Markers with LOD scores ≥2.7 and <3.5 identified four interactions, while those with ≤2.0 <2.7 LOD score identified three interactions and ≥3.5 identified 10 interactions. We only analyzed loci with ≥2.0 LOD score from all the chromosomes, therefore, there is a possibility that interactions of markers with <2.0 LOD score were missed. Indeed, we were surprised by the fact that some loci interacted at all the three time points, which included Sth4 on chromosome 4, Sth5 on chromosome 6, Sth8 on chromosome 9, and Sth10 on chromosome 13. This again shows that some genes are involved at all the three time points examined in this study. Therefore, epistatic interactions together with individual loci may constitute a genetic repertoire to promote wound healing.
We detected 17 epistatic interactions that could play a crucial role in wound healing. This study provides a wealth of information that should lead to more detailed understanding of wound healing. One unique feature of this study is the use of progenitor strains that have extreme phenotypes (SJL healed only 20% vs. 100% in MRL), therefore, the QTL in this study are expected to yield key wound-healing genes.
METHODS
Mice
The progenitor strains of MRL/MPJ females and SJL/J males were obtained from Jackson Laboratory. F1 from MRL/MPJ and SJL/J were crossed to get F2 population. These inbred strains of mice were selected from our earlier study of 20 different strains. The results indicated that MRL/MpJ completely heals and SJL/J heals <25% (Li et al. 2001b). Twenty, four-week-old female mice for MRL/MPJ and 10 SJL/J male mice were housed at the Animal Research Facility, J.L. Pettis Veterans Administration Medical Center, under the standard condition of 14 h light, 10 h darkness, ambient temperature of 20°C, and relative humidity of 30%–60%. All F1 and F2 mice (MRL/MPJ × SJL/J), crosses were bred at the Animal Research Facility. The experimental protocols were in compliance with the animal welfare regulation and approved by the J.L. Pettis Veterans Adminstration Medical Center.
Ear Punch and Measurement
Six hundred and thirty-three F2 female mice (MRL/MpJ × SJL/J) were generated to conduct the study. The 3-week-old F2animals had a 2-mm through-and-through hole in diameter made in the lower cartilaginous part of each ear using a metal ear punch (Fisher Scientific). The hole closure was measured using a 7× magnifier at days 15, 21, and 25 after ear punch. Each hole was measured twice (horizontal and vertical measurement) from the back of the ear (less hair at the back), and the average value for each animal was calculated from four measurements of two ears and was used in the data analysis. The precision of hole measurement, determined by repeatedly measuring the same hole 10 times, was 2.4% when the average size was 1.4 mm in diameter and 4.6% when the average size was 0.96 mm in diameter (Li et al. 2001b).
Genetic Analysis
The (MRL/MpJ × SJL/J) F2 population was killed at the age of 7 wk. Genomic DNA was prepared from the liver of each F2 animal population using a Wizard Genomic DNA kit (Promega), according to the manufacturer's instructions. DNA was stored at −35°C for later use. DNA was diluted 2:40 in 96 well plates before use for genotyping. Polymerase chain reaction (PCR) primers were purchased from Research Genetics to perform a genome-wide scan of the F2 mice population. Amplification was performed using Promega reagents (Promega). PCR cycling conditions included 4 min at 94°C × 1; 45 sec at 94°C; 30 sec at 52°C × 14; 45 sec at 94°C, 30 sec at 50°C, 1 min at 72°C × 32; 7 min at 72°C × 1. PCR products were resolved on 6% polyacrylamide gels (19:1 acrylamide: bis), stained with ethidium bromide, and visualized by ChemiImager 4400 Low Light Imaging System (Alpha Innotech Corporation). Pictures of the gels were taken and alleles recorded. Alleles derived from the MRL/MpJ parent were designated A, SJL/J derived alleles designated B, and H for heterozygotes, in data analysis.
Statistics
Genotype data was analyzed using a MAPQTL (4.0) program (CPRO-DLO). MAPQTLS interval mapping was utilized for quantitative trait loci mapping, and the LOD score significance thresholds were calculated using MAPQTL permutation test. A significant LOD score is ≥3.5. The significant LOD score for our data is the same as the values proposed by Van Ooijen (1999). A two-way ANOVA genome-wide scan was performed to identify epistatic interactions for markers with ≥ 2.0 LOD scores.
Acknowledgments
Assistance Award no. DAMD17–99–1–9571 supported this work. The US Army Medical Research Acquisition Activity is the awarding and administering acquisition office. The information contained in this paper does not necessarily reflect the position or policy of the Government and no official endorsement should be inferred. The authors thank the J.L. Pettis Veterans Administration Medical Center staff for their support. The authors also thank Melanie Hamilton-Ulland for excellent technical support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵1 Corresponding author.
-
E-MAIL Baylid{at}lom.med.va.gov; FAX (909) 796-1680.
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.203701.
-
- Received July 2, 2001.
- Accepted September 19, 2001.
- Cold Spring Harbor Laboratory Press