SWATH-MS-based proteogenomic analysis reveals the involvement of alternative splicing in poplar upon lead stress
- Fu-Yuan Zhu1,2,6,
- Xin Chen1,2,6,
- Yu-Chen Song1,2,6,
- Lydia Pui Ying Lam3,
- Yuki Tobimatsu4,
- Bei Gao5,
- Mo-Xian Chen1,2 and
- Fu-Liang Cao1,2
- 1The Southern Modern Forestry Collaborative Innovation Center, State Key Laboratory of Tree Genetics and Breeding, the Key Lab of Tree Genetics and Biotechnology of Educational Department of China and the Key Lab of Tree Genetics and Silvicultural Sciences of Jiangsu Province, Nanjing Forestry University, Nanjing 210037, China;
- 2Key Laboratory of State Forestry and Grassland Administration on Subtropical Forest Biodiversity Conservation, College of Biology and the Environment, Nanjing Forestry University, Nanjing 210037, China;
- 3Center for Crossover Education, Graduate School of Engineering Science, Akita University, Akita City 010-8502, Akita, Japan;
- 4Research Institute for Sustainable Humanosphere, Kyoto University, Uji, Kyoto 611-0011, Japan;
- 5National Key Laboratory of Ecological Security and Sustainable Development in Arid Areas, Urumqi, Xinjiang 830054, China
-
↵6 These authors contributed equally to this work.
Abstract
Alternative splicing (AS) regulates gene expression and increases proteomic diversity for the fine tuning of stress responses in plants, but the exact mechanism through which AS functions in plant stress responses is not thoroughly understood. Here, we investigated how AS functions in poplar (Populus trichocarpa), a popular plant for bioremediation, in response to lead (Pb) stress. Using a proteogenomic analysis, we determine that Pb stress induced alterations in AS patterns that are characterized by an increased use of nonconventional splice sites and a higher abundance of Pb-responsive splicing factors (SFs) associated with Pb-responsive transcription factors. A strong Pb(II)-inducible chaperone protein, PtHSP70, that undergoes AS was further characterized. Overexpression of its two spliced isoforms, PtHSP70-AS1 and PtHSP70-AS2, in poplar and Arabidopsis significantly enhances the tolerance to Pb. Further characterization shows that both isoforms can directly bind to Pb(II), and PtHSP70-AS2 exhibits 10-fold higher binding capacities and a greater increase in expression under Pb stress, thereby reducing cellular toxicity through Pb(II) extrusion and conferring Pb tolerance. AS of PtHSP70 is found to be regulated by PtU1-70K, a Pb(II)-inducible core SF involved in 5′-splice site recognition. Because the same splicing pattern is also found in HSP70 orthologs in other plant species, AS of HSP70 may be a common regulatory mechanism to cope with Pb(II) toxicity. Overall, we have revealed a novel post-transcriptional machinery that mediates heavy metal tolerance in diverse plant species. Our findings offer new molecular targets and bioengineering strategies for phytoremediation and provide new insight for future directions in AS research.
Rapid industrialization, urbanization, intensive agriculture, and mining have caused widespread contamination of soil and aquifer with heavy metals (HMs). In plants, lead (Pb) toxicity induces morphophysiological and biochemical changes and disrupts normal functions from the cellular level to the organ level (Ashraf and Tang 2017), which manifest in the inhibition of plant growth and photosynthesis, inactivation or denaturation of proteins and enzymes, and induction of oxidative stress and associated cellular damage (Gupta et al. 2013). To cope with the adverse effects of HM stresses, plants have developed a range of cellular mechanisms for detoxification, such as metal binding at cell walls, metal efflux via pumps in the plasma membrane, metal chelation in the cytosol, metal compartmentalization in the vacuole, and induction of antioxidants. In recent years, the identification of genes involved in Pb tolerance has provided new insights into how plants cope with Pb stress (Zhu et al. 2016). However, the intricate molecular mechanisms underlying such HM tolerance in plants are not fully understood, particularly at the post-transcriptional level.
Poplar is a model woody plant with economic value and plays an important role in ecosystems globally. It has great potential for phytoremediation due to its strong Pb resistance, fast growth, deep rooting, and low impact on food chains (Chen et al. 2014). A recent review (Luo et al. 2021) summarizes the Pb(II) uptake capacity of seven poplar species; among these, Populus nigra displays the strongest Pb tolerance, and Populus deltoides accumulates the largest amount of Pb in its roots, suggesting that poplar has great potential for the phytostabilization and ecological restoration of Pb-polluted soils. Although numerous studies regarding Pb uptake, translocation, and detoxification in poplar have been conducted, these studies primarily focused on the physiological and biochemical aspects. Currently, knowledge on the dynamic proteomic and transcriptomic changes under Pb stress in poplar is notably lacking. Thus, a comprehensive understanding of these mechanisms, which holds promise to help improving the phytoremediation ability of poplar that might also be applicable to other plants, is needed.
In the transcriptomic landscape, alternative splicing (AS) is regarded as an important mechanism for increasing the coding capacity of genomes (James et al. 2012; Rühl et al. 2012). Its identification and characterization in plants are widely considered a milestone to understanding their post-transcriptional machinery (Gupta et al. 2004; Nagasaki et al. 2006; Gu and Guo 2007). In total, 60%–85% of coding genes in plant genomes undergo AS, as revealed by recent genomic and proteogenomic studies of model plant species such as Arabidopsis (Arabidopsis thaliana) (Zhu et al. 2017), soybean (Glycine max) (Shen et al. 2014), rice (Oryza sativa) (Chen et al. 2020a), and maize (Zea mays) (Thatcher et al. 2014). Such genome-wide AS-induced gene expression changes have been shown to extensively influence plant development and growth processes, such as flowering, ageing, and fruit ripening (Syed et al. 2012; Zhu et al. 2013; Khan et al. 2014; Gupta et al. 2015; van Veen et al. 2016). Moreover, AS also plays an indispensable role in plant responses to abiotic stresses, such as flooding, drought, high salinity, and temperature stresses (Li et al. 2020b; Butt et al. 2022).
Although stress-induced genome-wide AS changes have been extensively documented in various plant species, the functional characterization of alternatively spliced isoforms has seldom been reported. In this study, to compare the spliced isoforms in poplar between the presence and absence of Pb(II), we applied parallel RNA sequencing (RNA-seq), including short-read RNA sequencing (srRNA-seq) and long-read RNA sequencing (lrRNA-seq), and a sequential window acquisition of all theoretical mass spectra (SWATH-MS)-based proteomic approach (i.e., proteogenomics). Transcriptomic and proteomic analyses have been individually applied in plant research to study various developmental processes and stress responses; however, these techniques are restricted by their shortcomings with respect to multivariate conditions and analytical pipelines. To solve this difficult problem, proteogenomics has emerged as an efficient method that can incorporate transcriptomic and proteomic data and represents a new generation of analytical approaches that can provide a more in-depth understanding of the importance of the potential genome coding ability (Castellana et al. 2014; Kumar et al. 2016). First, this analytical approach allows us to determine the wealth of information available at the proteomic level and apply it to assess the currently available genomic information of organisms, including identifying novel genes and spliced isoforms, assigning correct start sites and validating predicted exons and genes. Second, the use of SWATH-MS-based quantitative proteomics in proteogenomic analysis links the information of proteins to their transcript changes to accurately determine the protein abundance for each transcript isoform (Shen et al. 2021). This strategy could also greatly improve the weak correlations between protein abundance and transcript expression, facilitating further identification of valuable targets that are synchronously regulated at the transcript and protein levels.
In this study, we employed integrated proteogenomic analysis to analyze AS in poplar under Pb stress and explore the underlying AS mechanism. We further characterized how AS-regulated PtHSP70 mediates Pb resistance in poplar.
Results
Identification of diverse post-transcriptional events
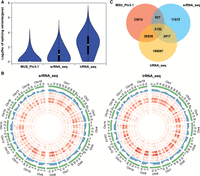
To determine the most effective method for identifying post-transcriptional events in response to Pb stress, the outputs of two different RNA sequencing approaches (srRNA-seq and lrRNA-seq) and the poplar genome annotation data available in a genome database (Phytozome v13) were compared. Compared with the srRNA-seq and poplar genome annotation methods, lrRNA-seq revealed the most diverse and abundant transcript isoforms (Fig. 1A,C). As illustrated by Circos representation (Krzywinski et al. 2009), the frequency and density of post-transcriptional events across the whole genome identified by lrRNA-seq were higher than those identified by srRNA-seq, suggesting that lrRNA-seq yielded higher transcript abundance and diversity (Fig. 1B). In addition, lrRNA-seq was previously shown to be a feasible approach for identifying longer transcripts, particularly for identifying post-transcriptional events (Chen et al. 2020b).
Comparison of transcript properties revealed by srRNA-seq and lrRNA-seq of Nanlin895 poplar plants (P. deltoides × P. euramericana). (A) Violin plot of spliced isoforms identified in the MSU_Ptv3.1 annotation, srRNA-seq, and lrRNA-seq data. (B) Circos representations (drawn by Circos) of post-transcriptional events identified from the srRNA-seq and lrRNA-seq data and abundance of transcripts recorded by MSU_Ptv3.1 annotation. The numbers correspond to the different layers of the Circos plot and are ordered from outwards to inwards. 1, intron retention (IR); 2, multi-IR (MIR); 3, skipped exon (SKIP); 4, multiexon SKIP (MSKIP); 5, alternative exon 5′ (AE5′); 6, alternative exon 3′ (AE3′); 7, alternative transcription start (ATS); 8, alternative polyadenylation (APA); 9, alternative 5′ first exon (AFE); 10, alternative 3′ last exon (ALE). (C) Venn diagram showing the overlapping transcripts identified from the MSU_Ptv3.1 annotation, srRNA-seq, and lrRNA-seq data.
Deep RNA-seq reveals extensive AS in response to Pb treatment
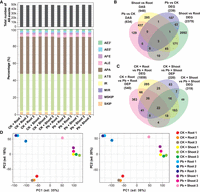
To determine AS in poplar in response to Pb stress and validate the integrity and veracity of our transcriptome data, we first performed a transcriptomic analysis to identify AS events in poplar seedlings treated with or without Pb(NO3)2 (0.75 mM) for 24 h as described previously (Zhu et al. 2016) via both srRNA-seq and lrRNA-seq. We divided the poplar seedlings into roots and shoots due to the difference in the mRNA expression levels of the response genes in these two parts. Approximately 50,000 post-transcriptional events were identified in each sample. Among these, alternative polyadenylation (APA) and alternative transcription start (ATS) sites accounted for more than 80% of the total events identified in all the samples (Fig. 2A). As expected, alternative first exons (AFEs) and alternative last exons (ALEs) were the two most abundant types of AS events (the pattern diagram for identifying AS events is provided in Supplemental Fig. S9) in all the samples (Fig. 2A). A comparison of the identified differentially expressed genes (DEGs) (Supplemental Table S1) with the genes undergoing differential AS (DAS) (Supplemental Table S2), which were obtained with the Pacific Biosciences (PacBio) RSH platforms, revealed that more than 94% of the data did not overlap. Only 241 genes showed differential expression at both the transcriptional and post-transcriptional levels (Fig. 2B), indicating that AS may play a role distinct from transcriptional regulation in poplar in response to Pb stress. Another comparison was conducted between the DEGs and differentially expressed proteins (DEPs) identified from SWATH-MS-based proteomic data. Our data suggested that 335 genes were shared among the sets of DEGs and DEPs (Fig. 2C). Preliminary data exploration with principal component analysis (PCA) was conducted using the cleaned sequencing read data to assess the factors that contributed most to the observed transcriptional variation between samples. The replicates of each treatment-tissue combination clustered closely together, indicating that the RNA-seq data were reliable (Fig. 2D).
Bioinformatic comparison of the data sets of DEGs and genes subjected to DAS. (A) Summary of the total number and types of AS events identified. Data from each biological replicate are shown. (B,C) Venn diagrams displaying the common and unique genes between the data sets of DEGs and genes subjected to DAS and between the data sets of DEPs and DEGs. (D) Principal component analysis (PCA) of all 12 samples, which were divided into four groups. ALE, alternative last exon; AFE, alternative first exon; SKIP, exon skipping; MSKIP, multiple exon skipping; MIR, multiple intron retention; IR, intron retention. Group 1, CK shoots versus CK roots; Group 2, Pb roots versus CK roots; Group 3, Pb shoots versus CK shoots; Group 4, Pb shoots versus Pb roots; CK, control sample grown in the absence of Pb; Pb, sample grown in the presence of Pb.
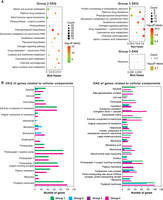
We further performed a Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis to determine whether these DEGs were significantly enriched in specific functions. The majority of the pathways enriched with the DEGs were closely related to cellular metabolic processes, such as phenylpropanoid biosynthesis, glutathione metabolism, and starch and sucrose metabolism, suggesting the potential contribution of these pathways to Pb detoxification (Fig. 3A). In addition, a Gene Ontology (GO) pathway analysis of the DEGs and genes that undergo DAS upon Pb exposure was conducted to provide a quick overview of the significant pathways. Different pathways related to cellular components, particularly the intracellular, membrane, and microtubule pathways, were significantly enriched with DEGs and the genes underwent DAS in the roots under Pb treatment (Fig. 3B), suggesting that both DEGs and DAS mediated a number of cellular pathways in response to Pb stress. Thus, cell structures such as membranes and microtubules may play a role in the responses to Pb stress (Supplemental Fig. S1B). Overall, our proteogenomic data demonstrated that AS occurs extensively in poplar in response to Pb stress.
Enrichment analysis of DEGs and genes subjected to DAS events. (A) KEGG pathway enrichment analysis of the DEGs and genes subjected to DAS events in Group 2 and Group 3. (B) GO enrichment analysis of genes enriched in cellular components between the data sets of DEGs and genes subjected to DAS. Group 1, CK shoots versus CK roots; Group 2, Pb roots versus CK roots; Group 3, Pb shoots versus CK shoots; Group 4, Pb shoots versus Pb roots; CK, control sample grown in the absence of Pb; Pb, sample grown in the presence of Pb.
Nonconventional splice site utilization is higher in poplar seedlings under Pb stress
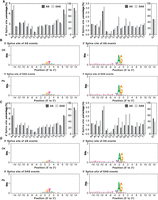
To further understand the role of AS in response to Pb stress in poplar seedlings, the splice site usage was analyzed by quantifying the frequency of splicing site selection and conducting a conservation analysis of the splice sites (Fig. 4). Previous studies have suggested that conventional U2-type splice sites (5′-GT–AG-3′) are conserved and account for ∼90% of total splice sites among plant species (Chen et al. 2019). Consistent with the previous findings, these conventional splice sites accounted for 50% and 87% of the 5′ and 3′ splice sites, respectively, in the control roots (Fig. 4A). These splice sites decreased to 46% and 79% under Pb treatment, and conversely, the percentages of nonconventional AS sites reached 110%–200% relative to those identified in the controls (Fig. 4A), suggesting that some of the DAS events observed under Pb treatment resulted from the increased use of nonconventional splice sites. Importantly, SNPs between P. trichocarpa and (Populus deltoides × Populus euramericana) did not affect the uses of nonconventional splicing sites under Pb(II) stress according to our genomic comparison analysis (Supplemental Fig. S4C). A subsequent conservation analysis revealed that the 3′-splice sites in poplar seedlings were characterized by a “CAG” signature, and Pb treatment did not change this signature (Fig. 4B). Similar results were found in shoots (Fig. 4C,D). The “GGT” signature was also identified in both the AS and DAS data sets. Although almost all the genes showed different expression levels in the control roots and shoots, their uses of splice sites were similar. In contrast, under Pb treatment, the uses of nonconventional AS sites, including intron retention (IR), AFEs and ALEs, were increased in the shoots but not in the roots (Supplemental Fig. S2). Accordingly, poplar probably adapts to Pb stress through the modulation of RNA splicing with the uses of nonconventional splice sites, yielding novel spliced transcripts that could contribute to Pb detoxification.
Frequency of splice site selection under Pb stress. (A,B) Frequency of splice site selection in total AS events and Pb-affected DAS events and frequency of splice site selection in total AS events and Pb-affected DAS events in Group 2. (C,D) Frequency of splice site selection in total AS events and Pb-affected DAS events and frequency of splice site selection in total AS events and Pb-affected DAS events in Group 3. Group 1, CK shoots versus CK roots; Group 2, Pb roots versus CK roots; Group 3, Pb shoots versus CK shoots; Group 4, Pb shoots versus Pb roots; CK, control sample grown in the absence of Pb; Pb, sample grown in the presence of Pb.
Transcription factor (TF)–SF synchronization in poplar in response to Pb stress
Splicing factors (SFs) and TFs are typical factors that regulate AS. SF directly mediates the AS pattern, whereas TF influences pre-mRNA synthesis, including RNA polymerase II (RNAPII) elongation, which is known to impact cotranscriptional pre-mRNA processing such as splicing (Rambout et al. 2018).
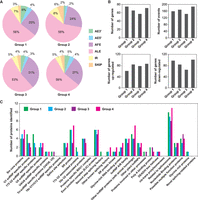
To identify the specific factors that regulate AS under Pb stress, Pb(II)-responsive SFs and TFs in poplar were analyzed with respect to their AS patterns and expression levels. Sixty SF genes were found to undergo DAS, and a total of 120 AS events were detected (Fig. 5). Among these, 60 AS events showed an increase in frequency, whereas the frequency of the other 60 AS events was decreased (Fig. 5B). In detail, ∼83% of the identified AS events were AFEs and ALEs (Fig. 5A). These SFs were further classified into 29 subgroups via our curated SF database reported previously (PlantSPEAD, http://chemyang.ccnu.edu.cn/ccb/database/PlantSPEAD/) (Chen et al. 2021). These include Sm core proteins related to spliceosomes, SR proteins, and proteins related to splice site selection (Fig. 5C), indicating that conventional SFs may also play roles in the responses to Pb stress.
Splicing event statistics and groups of SFs. (A) Proportion of splicing events in each compared group. (B) Numbers of identified proteins in each group. (C) Numbers of genes, events, increased events, and decreased events. Group 1, CK shoots versus CK roots; Group 2, Pb roots versus CK roots; Group 3, Pb shoots versus CK shoots; Group 4, Pb shoots versus Pb roots; CK, control sample grown in the absence of Pb; Pb, sample grown in the presence of Pb.
TFs are important regulators that regulate gene expression in response to environmental stresses, such as drought, temperature, salt, and UV radiation (Li et al. 2020b). In our root and shoot DAS data sets, more than 290 TFs categorized in 35 TF families were found (Supplemental Fig. S3), suggesting that TFs undergo substantial post-transcriptional regulation in response to Pb stress. Among these families, C2H2s (zinc finger proteins), ERFs (ethylene-responsive factor), and WRKYs (WRKY transcription factors) accounted for a large proportion (∼40%), which is suggestive of their potential involvement in Pb responses. In all these TFs, the most prevalent AS events were AFEs and ALEs (72%) (Supplemental Fig. S3A).
To further investigate the relationship between TFs and SFs associated with AS during Pb stress, we integrated transcriptomic and proteomic approaches to systematically reveal multiple layers of regulatory networks that function during Pb stress. By overlapping the poplar SFs and TFs in the DAS data set, we selected SFs and TFs belonging to Group 2 (Pb-treated roots vs. H2O-control roots) and Group 3 (Pb-treated shoots vs. H2O-control shoots) and constructed a heatmap to illustrate the differential expression of SF- and TF-encoding genes that undergo AS (Supplemental Fig. S4). Based on previous publications on plant tolerance to HMs, we constructed an HM-related gene list that includes 89 genes (Supplemental Table S3). We compared this list with the SF- and TF-encoding genes in the DAS data set and found several overlapping candidate genes that could be involved in TF–SF synchronization in the HM tolerance response. We also found that they are Pb(II)-inducible (Supplemental Fig. S4), which would certainly indicate their potential as targets in molecular breeding of poplar for HM resistance by further functional investigations.
HSP70 plays an important role in Pb tolerance
To further identify critical genes contributing to Pb tolerance, a comprehensive bioinformatics analysis coupled with functional characterization was conducted. Our data indicated that thousands of genes produced multiple transcript isoforms and that hundreds of proteins were differentially expressed in response to Pb treatment (Supplemental Tables S1, S2). We selected four genes that were classified as both DEGs and DEPs in roots to verify their expression by RT–qPCR (Supplemental Fig. S5B). Consistent with the transcriptomic data, the expression of these genes was strongly up-regulated in both roots and shoots upon Pb exposure. From this set of genes, we chose a HSP70 family gene (PtHSP70; Potri.010G205800) whose two isoforms (PtHSP70-AS1 and PtHSP70-AS2; Potri.010G205800.1 and Potri.010G205800.2) showed increased expression in both the roots and the shoots under Pb treatment for further functional characterization. Such Pb(II)-inducible expression of PtHSP70 was also observed in the four other commonly used poplar species (Supplemental Fig. S5C).
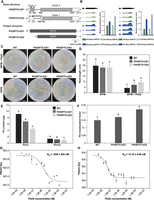
The two spliced isoforms of PtHSP70 are generated by the 5′ alternative exon (5′AE) (Fig. 6A). PtHSP70-AS1 is composed of two exons interrupted by an intron. In contrast, the translation of PtHSP70-AS2 starts from the first exon. Thus, the protein encoded by PtHSP70-AS2 is shorter than that encoded by PtHSP70-AS1 by 105 amino acids at its N-terminus. The wiggle plots (drawn by the Integrative Genomics Viewer) (Thorvaldsdóttir et al. 2013) based on RNA-seq for PtHSP70 are shown in Figure 6B.
Gene and protein structure and expression of PtHSP70, and phenotypic analysis of transgenic poplar overexpressing PtHSP70-AS1 and PtHSP70-AS2. (A) Gene and protein structures of PtHSP70-AS1 and PtHSP70-AS2. Dotted line, the reverse primer of PtHSP70-AS2 is a splicing junction primer that span the intron. The reverse primer of PtHSP70-AS1 starts at the exon which PtHSP70-AS2 does not have. (B) RNA-seq read mapping data for RefSeq annotated gene regions for PtHSP70 and the expression of PtHSP70-AS1 and PtHSP70-AS2 in wild-type poplar treated and not treated with Pb. (C) Phenotype of transgenic poplar overexpressing PtHSP70-AS1 and PtHSP70-AS2 after Pb treatment. (D) Root length of transgenic poplar overexpressing PtHSP70-AS1 and PtHSP70-AS2 after Pb treatment. (E) Pb contents of transgenic poplar overexpressing PtHSP70-AS1 and PtHSP70-AS2 after Pb treatment. (F) Pb translocation factors of transgenic poplar overexpressing PtHSP70-AS1 and PtHSP70-AS2 after Pb treatment. The letters represent significant differences between the WT and transgenic lines (ANOVA, P < 0.05). CK, control sample grown in the absence of Pb; Pb, sample grown in the presence of Pb; WT, wild type; PtHSP70-AS1, transgenic poplar overexpressing PtHSP70-AS1; PtHSP70-AS2-1, transgenic poplar overexpressing PtHSP70-AS2. (G,H) Binding affinity between Pb(II) and recombinant PtHSP70-AS1 (G) and PtHSP70-AS2 (H) determined by MST binding assay. The values represent the means ± SDs (n = 3).
To understand the function of HSP70 in the Pb response, we generated transgenic poplar and Arabidopsis plants overexpressing PtHSP70-AS1 or PtHSP70-AS2 and cultivated them side-by-side with wild-type (WT) controls on media with or without Pb(NO3)2. We measured and compared their root length, which is a typical indicator of the Pb tolerance of plants (Yu et al. 2012). In the absence of Pb treatment, the root length of the transgenic poplar plants overexpressing either PtHSP70-AS1 or PtHSP70-AS2 was similar to that of the WT controls (Fig. 6D). However, under Pb treatment, the root length of the transgenic poplar was significantly longer than that of the WT controls. A similar observation was also found with transgenic Arabidopsis overexpressing PtHSP70-AS1 or PtHSP70-AS2 (Supplemental Fig. S6B). In contrast, the phenotype of empty vector (EV) line under Pb(II) treatment was similar to WT plants (Supplemental Fig. S6C). Taken together, the results indicate that the overexpression of both spliced isoforms of PtHSP70 confers Pb tolerance in poplar and Arabidopsis, supporting our view that the spliced isoforms of HSP70 help plants to resist Pb toxicity. We subsequently further investigated the underlying mechanism and determined the SF that regulates AS of PtHSP70.
Exploration of the mechanisms of HSP70 tolerance to Pb
We further investigated how the two splicing isoforms of PtHSP70 function in Pb tolerance in poplar. First, we determined the capacities for Pb(II) translocation from the roots to the shoots in transgenic poplar overexpressing PtHSP70-AS1 or PtHSP70-AS2. Measurements of the Pb(II) contents in these poplars revealed that both the roots and shoots of the transgenic poplar plants overexpressing either PtHSP70-AS1 or PtHSP70-AS2 contained less Pb(II) than those of the WT controls (Fig. 6E). Furthermore, the translocation factors, which are defined by the ratio of the HM content in the aboveground part (shoots) to that in the belowground part (roots) of a plant (Cheng et al. 2021), were higher in the transgenic poplar than in the WT controls (Fig. 6F), suggesting that the transgenic poplar lines exhibit stronger capacities for Pb(II) translocation from the roots to the shoots. These results suggest that the two PtHSP70 isoforms participate in regulating the reduction in cellular toxicity in poplar through Pb(II) extrusion.
Considering the chaperone activities and transport capacity of HSP70, we hypothesized that HSP70 may function as an HM storage protein to cope with Pb stress. Thus, we sought to investigate how HSP70 promotes Pb tolerance in plants by microscale thermophoresis (MST), which is a novel technique to detect protein-molecule interactions. To examine the direct interaction of HSP70 proteins with Pb(II), bacterially expressed recombinant PtHSP70-AS1 and PtHSP70-AS2 proteins were incubated together with Pb(II) at different concentrations and subjected to the MST-based binding assay. Our results showed that both PtHSP70-AS1 and PtHSP70-AS2 could spontaneously bind to Pb(II) (Fig. 6G,H). Notably, the binding affinity of PtHSP70-AS2 (Kd = 15.7 ± 9.86 nM) was more than 10-fold higher than that of PtHSP70-AS1 (Kd = 1659 ± 803 nM). Furthermore, the notably higher binding capacity of PtHSP70-AS2 compared with that of PtHSP70-AS1 is in line with the stronger tolerance of the PtHSP70-AS2-overexpressing poplar plants compared with that of the PtHSP70-AS1-overexpressing poplar plants during Pb(II) treatment (Fig. 6C–F). The above-described results indicate that the spliced isoforms of PtHSP70 confer Pb tolerance through direct binding to Pb(II) and reducing cellular toxicity through Pb(II) extrusion.
We further identified the SF that regulates AS of PtHSP70. We previously demonstrated that the core SF U1-70K plays a crucial role in regulating AS in plants (Chen et al. 2020b). Here, using transgenic poplar overexpressing PtU1-70k, we found that the overexpression of PtU1-70k increased the expression of both PtHSP70-AS1 and PtHSP70-AS2 (Supplemental Fig. S7B), supporting the notion that a potential AS regulation by U1-70K-HSP70 contributes to Pb tolerance in poplar. Other Pb(II)-responsive SFs belonging to the U1 complex, such as U1C and Luc7, were also identified by our proteogenomic analysis (Supplemental Table S2), implying that these SF-associated U1 modules reciprocally regulate the AS of PtHSP70 to generate more spliced transcripts to confer Pb resistance in poplar.
Conservation of AS of HSP70 orthologs in plants
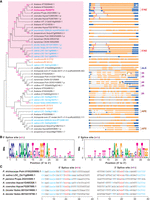
To elucidate the involvement of HSP70 in plant tolerance to HM stress via AS, we analyzed the AS patterns of HSP70 orthologs across the plant kingdom. A BLASTN search of these HSP70s retrieved a total of 46 sequences of the AS isoforms of HSP70 orthologs from 14 plant species, including 10 eudicots, three monocots, and one gymnosperm (Fig. 7A). Using the sequences of the longest proteins encoded by these isoforms, we constructed a phylogenetic tree to interpret their phylogenetic relationship. The protein sequence was identified with NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Based on the branching pattern of the phylogenetic tree and protein sequence identity with PtHSP70, we defined 18 proteins in the same clade as the orthologs of PtHSP70 (highlighted in pink in Fig. 7A). Through the tree, we found that the same AS events are often occurred in HSP70 orthologs. This indicates that AS patterns may be evolutionarily conserved, and the conserved sequences of the splicing sites or AS events are potential predictors of gene function. An analysis of their AS patterns suggested that a rice ortholog (LOC_Os11g08460.1) also undergoes the same 5′AE as PtHSP70. Further analysis of the flanking sequences of the potential splicing sites of all orthologs by WebLogo and multiple sequence alignment revealed that the other 5 orthologs harbor the same 5′ and 3′ splicing sites, although the corresponding spliced isoforms were not found in the databases (Fig. 7B,C). In contrast, the genes encoding proteins distantly related to PtHSP70 experience different types of AS events, including IR and AFE/ALE. Collectively, the 5′AE-driven splicing machinery on PtHSP70 was also found in its orthologs, indicating the possible existence of a common regulome during Pb(II) toxicity in plants.
Phylogenetic and splice site analysis of HSP70. (A) Phylogenetic tree of PtHSP70 proteins. The unrooted phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replications. Blue, monocots; black, eudicots; orange, gymnosperm. The structure of each AS isoform of HSP70 genes and its corresponding splicing event are shown. Pink arrows, 5′ splice sites; purple arrows, 3′ splice sites; solid arrows, same splice sites as PtHSP70 in which AS occurs; open arrows, same splice sites as PtHSP70 but in which AS does not occur; 5′AE, 5′ alternative exon; ALE, alternative last exon; AFE, alternative first exon; IR, intron retention. (B,C) Analysis of splice site sequences of HSP70 genes. The flanking sequences of splice sites of HSP70 (Fig. 7A) were analyzed by WebLogo (B) and multiple sequence alignment (C). The conserved sequences are highlighted in red and blue.
Discussion
AS occurs in poplar during Pb stress
AS is an important process to increase the genomic coding capacity of plants, and its involvement in plant tolerance and adaptation to various abiotic stresses, such as drought, high salinity, low oxygen, and extreme temperatures, has been extensively studied (Laloum et al. 2018). In the present study, an AS analysis revealed that most of the genes undergoing DAS showed no change in their expression levels, suggesting that the regulation of transcripts by AS is mostly separated from the transcriptional control of conventional DEGs (Fig. 2B). Under Pb stress, DEGs produced various transcripts mediated by different AS events. Among these events, AFEs and ALEs were the most abundant, which resulted in variable 5′ and 3′ untranslated ends and likely affected the efficiency of translation or the stability of the corresponding transcripts (Sonmez et al. 2011; Jenal et al. 2012). Further AS analysis indicated that under Pb stress, DAS of a number of genes occurred due to an increased use of nonconventional splice sites, which are particularly prevalent in some AS events, such as AFEs, ALEs, and IR. Therefore, AS is probably involved in the Pb stress response in poplar via the recruitment of nonconventional splice sites. Moreover, SFs were subjected to AS mediated by AFE and ALE events (Fig. 5A). Thus, these events constitute the core components of the mechanism through which AS is involved in the response to Pb stress. We quantitively identified a number of Pb-responsive SFs, such as the components of the U1 complex, which itself is a component of the spliceosome (Chen et al. 2020b). Our results indicated that the Pb-induced transcriptome of poplar is subjected to substantial AS mediated by the potential spliceosome. Furthermore, the numbers of TF- and SF-encoding genes subjected to AS were similar (Supplemental Fig. S3). Emerging evidence has revealed that a massive and rapid AS response occurs during exposure to cold stress and regulates cold-responsive TFs and SFs/RNA-binding proteins (Li et al. 2020a). For example, recent studies have shown that the AS of TFs generates small interfering peptides that negatively constitute self-regulatory circuits in plants under cold stress. Moreover, a number of SFs involved in RNA binding, splice site selection, and spliceosome assembly are also affected by temperature fluctuations (Seo et al. 2013). These findings support a close association between TFs and SFs in response to abiotic stress. Here, our analysis showed that the SF- and TF-encoding genes that undergo AS produce different spliced isoforms and exhibit different expression levels under Pb stress (Supplemental Fig. S4). Additionally, a number of TFs associated with SFs are highly relevant to the HM response (Supplemental Fig. S4), and these findings provide additional support for the notion that TF–SF synchronization modulates Pb(II) detoxification in poplar.
In this study, we used a novel integrated approach that involves both transcriptomic and proteomic analyses to study the response of poplar to HMs. Such proteogenomics is considered a powerful tool for studying AS-involved biological processes (Blank-Landeshammer et al. 2019). Through this approach, the roles of AS in response to Pb stress were clarified, and many meaningful SFs and Pb-responsive genes were identified. The evidence further supports that proteogenomics is an effective method for future studies on the molecular mechanism and post-transcriptional regulation of plants in response to abiotic stress.
The AS of HSP70 confers Pb tolerance
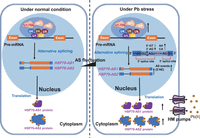
Our proteogenomic analysis revealed that a Pb(II)-inducible chaperone protein, PtHSP70, is regulated by AS in poplar under Pb stress. In general, HSP70 proteins serve as molecular chaperones, and their expression is markedly induced by adverse environmental stresses. The HSP70 family is highly conserved among prokaryotes and eukaryotes (Song et al. 2021). Under stress, HSP70 helps denatured proteins to refold, prevents denatured proteins from aggregating, and solubilizes or degrades protein aggregates (Leng et al. 2017). Previous studies have also found that HSP70 can serve as an ATP-binding cassette (ABC) transporter during HM stress responses (Kim et al. 2001). The functions of HSP70 in poplar under Pb(II) stress are intriguing, particularly with respect to the roles of HSP70 spliced isoforms. Our characterizations of transgenic poplar overexpressing PtHSP70 spliced isoforms as well as their recombinant proteins revealed that HSP70 may act as a transporter protein that directly carries Pb(II) out of cells, thereby preventing Pb(II)-induced cellular damage in poplar (Fig. 6). Both PtHSP70-AS1 and PtHSP70-AS2 show high affinity to Pb(II), but the two isoforms have different binding capacities. Specifically, PtHSP70-AS2 has a notably stronger binding affinity to Pb(II) than PtHSP70-AS1 (Fig. 6G,H). PtHSP70-AS2 is different from PtHSP70-AS1 by a truncation at the N-terminus (Fig. 6A). It is possible that this truncation removes some regions that interfere with Pb(II) binding and/or leads to the exposure of some internal regions that have higher binding affinity with Pb(II). A subcellular localization analysis also revealed that the shorter form of PtHSP70-AS2 prefers to localize in the cytoplasm (Supplemental Fig. S8C), thus facilitating the binding of Pb(II) to rapidly reduce intracellular toxicity. We also examined the potential interaction between PtHSP70-AS1 and PtHSP70-AS2 by yeast two-hybrid (Y2H), but no interaction was observed (Supplemental Fig. S8B), indicating that each of these proteins can resist Pb without relying on mutual interaction. Furthermore, a significant decline in the Pb(II) contents was observed in both the PtHSP70AS1- and PtHSP70AS2-overexpressing lines compared with the WT control, indicating that HSP70 alleviates poplar Pb(II) toxicity through extrusion (Fig. 6E). Overall, AS may increase the total number of spliced transcripts of PtHSP70 to prevent Pb(II) toxicity and reduce HM damage. In particular, one of the spliced isoforms of HSP70, that is, PtHSP70-AS2, may be specifically generated via AS to improve the capacity to transport or store Pb(II), thus reducing HM damage. Regarding the possible underlying mechanism, a recent study showed that HSP70 can interact with phospholipase D delta and participate in defence against heat stress in plants (Song et al. 2021). However, we found that PtHSP70 does not interact with PLDD (Supplemental Fig. S8A). Hence, we speculate that HSP70 may carry Pb(II) ions out of plant cells through interactions with some HM pumps (Fig. 8) that reportedly functions as a Pb(II) pump at the plasma membrane (Lee et al. 2005).
In summary, we have shown that AS is an important component of the HM detoxification mechanism in poplar. Furthermore, the AS of PtHSP70 confers Pb tolerance in poplar by generating spliced isoforms that are capable of binding to Pb(II) directly and reducing cellular toxicity through Pb(II) extrusion. Based on the identified list of proteins regulated by AS during Pb stress, future functional characterization of the other proteins regulated by AS and identification of the SFs that regulate such AS events would improve our understanding of how AS modulates the abiotic stress responses of plants. Such findings will aid the development of novel bioengineering approaches to improve the efficiency of phytoremediation and of new strategies to enhance the productivity of plants in unfavorable environments, such as those with HM contamination. Because the AS of HSP70 appears to be highly conserved in plants, this knowledge and bioengineering approaches may also be applicable to improve the HM tolerance of other plant species that also show great potential for phytoremediation.
Methods
Plant materials, growth conditions, and Pb treatment
Four- to five-week-old “Nanlin 895” (Populus deltoides × Populus euramericana) cultured seedlings were removed from woody plant medium (WPM) and transferred to bottles containing water or treatment solution supplemented with 0.75 mM Pb(NO3)2. Pb(II) concentration was selected based on the results of our pretreatment experiments and literature (Szuba et al. 2020). In our pretreatment experiments, different concentrations of Pb(II) were used to treat poplar seedlings, and 0.75 mM were selected as the appropriate concentrations according to the inhibition rate of root growth. The seedlings were incubated in a culture room at 23°C with a light/dark cycle of 16/8 h for 24 h. For further transcriptomic and proteomic analyses, three biological replicates of Pb-treated and untreated poplar seedlings were used. After treatment, the plants were divided into two parts: the ground part (shoot) and the lower part (root).
Seeds of Arabidopsis (Col-0) were surface-sterilized with 20% (v/v) NaClO and 75% (v/v) ethanol and then sown on ½ Murashige and Skoog medium plates (Murashige and Skoog 1962) supplemented with 0.8% (w/v) agar and 1.5% (w/v) sucrose. After 2 d of stratification, the plates were incubated under 16-h light (23°C)/8-h dark (21°C) cycles.
Generation of transgenic plants
To generate PtHSP70-AS1- and PtHSP70-AS2-overexpressing Arabidopsis and poplar (Populus deltoides × Populus euramericana), full-length coding sequences of PtHSP70-AS1 and PtHSP70-AS2 were cloned under the control of a cauliflower mosaic virus 35S promoter in a pMDC43 binary vector. The final constructs were transformed into WT Arabidopsis (Col-0) by the floral dip method using Agrobacterium tumefaciens strain GV3101 (Clough and Bent 1998). The transformation of poplar was conducted as previously described (Neb et al. 2017). Transgenic Arabidopsis was cultivated on ½MS plates (Islam et al. 2021), and transgenic poplar plants were grown on WPM medium with MES and Ca2+ under the above-described growth conditions.
Assays of Pb(II)-induced root inhibition in Arabidopsis and poplar
For the phenotypic comparison of Arabidopsis, the seedlings were planted on ½MS board for 3–4 d and then transferred to another board, either ½MS board (denoted ½MS) or ½MS board with 0.5 mM Pb(NO3)2 (denoted ½MS + Pb) for 7 d. For the phenotypic comparison of poplar, 5-wk-old seedlings were transferred to WPM or WPM with 1 mM Pb(NO3)2 (denoted WPM + Pb) for 2 mo. The root length was measured, and physiological data were calculated for the quantification of Pb(II) tolerance.
Plant RNA extraction and srRNA-seq
For the extraction of total RNA, shoots and roots of poplar were ground in liquid nitrogen and extracted using an RNAprep Pure Plant Plus Kit (Polysaccharides & Polyphenolics-rich). The extracted RNA was qualified using a LifeReal spectrophotometer (NanoReady). srRNA-seq experiments were conducted as previously described with minor modifications (Zhu et al. 2017). The resulting cDNA library constructed from poplar RNA samples was used for paired-end (2 × 125-bp) sequencing with an Illumina HiSeq 4000 platform by Annogroad Gene Technology Co., Ltd. (Beijing, China). Three replicates of each sample were trimmed to obtain clean reads for subsequent analysis.
Single-molecule lrRNA-seq and data analysis
The library construction steps and sequencing strategies were described previously (Zhu et al. 2017) and performed with minor modifications. In general, five libraries (i.e., 0.5–1 k, 1–2 k, 2–3 k, 3–6 k, and 5–10 k) were generated and sequenced using 16 SMRT cells for each tissue type on a PacBio RSII platform (BGI). The resulting raw data were processed by the ToFU pipeline as described on the company website (Zhu et al. 2017). Both high- and low-quality full-length transcripts were subjected to base correction by two rounds of BLAST searches against the Phytozome reference genome and cDNA sequences for subsequent bioinformatic analysis.
Analysis of RNA-seq and proteomic data
The P. trichocarpa reference genome annotation file was downloaded from Phytozome v13 (https://phytozome-next.jgi.doe.gov/). The mapping of clean reads and subsequent bioinformatic analysis were performed as described previously (Zhu et al. 2017). The analytical pipeline was summarized previously (Zhu et al. 2017), and significant changes in differentially expressed genes (DEGs) (Supplemental Table S1) and differentially expressed alternative splicing (DAS) events (Supplemental Table S2) were determined by log2-fold changes (log2FC) > 2 and Q-value (FDR < 5%).
Transcript remapping and identification of AS
The P. trichocarpa genome sequences were downloaded from Phytozome v13 (https://phytozome-next.jgi.doe.gov/). Remapping of the previously genome-guided assembled transcripts (total 22168) from Illumina stranded paired-end reads (srRNA-seq data set) and PacBio full-length transcripts (total 228765) from the lrRNA-seq dataset to the poplar genome was performed using GMAP (Abdel-Ghany et al. 2016).
Further filtering was performed by comparison with the extant poplar gene models, and transcripts that contained at least one correct junction or covered an intact exon were retained. AS events were then analyzed using ASprofile (https://ccb.jhu.edu/software/ASprofile/) as previously described (Zhu et al. 2017). A Circos diagram was drafted using the AS frequency mapped to the poplar genome. Additionally, the splice site statistics and conservation analysis were summarized and constructed using the online software WebLogo v3 (https://weblogo.threeplusone.com) (Crooks et al. 2004). Splicing variants were identified using full-length transcripts after two rounds of correction against the P. trichocarpa reference genome and cDNAs.
SWATH-MS analysis
SWATH methods were used for subsequent MS analysis using a TripleTOF 5600 (Sciex) LC/MS system. The prepared samples were bound to the trap column and separated using an analytical column (45-min gradient, total time of 60 min) and a 120-min linear gradient of two mobile phases (buffer A [0.1% (v/v) formic acid, 5% (v/v) DMSO in water] and buffer B [0.1% (v/v) formic acid, 5% (v/v) DMSO in acetonitrile]) from 5% to 35% buffer B at 300 nL/min. A survey scan of each cycle was performed to obtain the 250-ms MS1 scan in the range of 350–1500 m/z and 250-ms MS2 scan in the range of 350–1500 m/z over a 100-sheet variable window.
The mass spectrometry file obtained from the SWATH scan was processed by DIA-Umpire to obtain the secondary mass spectrometry file that can be used for database search. The Comet and X!Tandem search engines in TPP software were used for the database searches. The search results were used as a spectrum library for subsequent targeted extraction. Open SWATH was used as the algorithm for SWATH-targeted extraction and quantification (Rost et al. 2014). The test results were screened based on a false discovery rate (FDR) of 1%. The protein quantitative intensity information obtained from the SWATH analysis was subjected to log2 conversion, data filling, and data normalization using the imputation algorithm in Perseus software for difference comparison and t-test analysis. Proteins of shoots and roots with a ratio higher than 5 or lower than 0.2 (P < 0.05) were considered DEPs in this study. The spectrum information of SWATH-MS can be seen in Supplemental Figure S10. The results of lrRNA-seq were translated into protein sequences for SWATH-based database comparison, and this information have been added to the Supplemental Files (Supplemental Table S5).
Quantitative RT–PCR (RT–qPCR) validation of AS transcripts
Total RNA (∼5 μg) was reverse-transcribed into cDNA using the SuperScript First-Strand Synthesis System (Invitrogen) following the manufacturer's instructions. RT–qPCR was conducted as described previously (Zhu et al. 2013) and based on previous standard rules established in plant research (Udvardi et al. 2008). PCR was performed with a StepOne Plus Real-time PCR System (Applied Biosystems) using SYBR Green Pro Taq Premix (Accurate Biology). The program used was as follows: 95°C for 30 sec followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. Three biological replicates were used in all RT–qPCR experiments. PtUBIC (ubiquitin-conjugating) and PtACT6 (Actin6) were used as internal reference genes (Supplemental Fig. S11). All the primers, including the isoform-specific primers used for the RT–qPCR experiments, are listed in Supplemental Table S4.
Measurement of the Pb(II) content in poplar
Transgenic PtHSP70-overexpressing poplar lines were treated with H2O (control) or 1 mM Pb(NO3)2 (five repetitions of each treatment). After 2 mo of treatment, the growth of the plants was observed, and the root length and plant height were measured. We then divided the plants into root and shoot parts, dried these parts for 12 h in an oven at 55°C and ground them into powder. The concentrations of Pb were measured according to the method described by Cheng et al. (2021). Briefly, sample powder (0.2g) was digested with an acid mixture (HNO3/HClO4, v/v = 4/1) at 250°C for 3 h, diluted to 25 mL, and filtered. The concentrations of metals were determined by inductively coupled plasma-mass spectrometry (ICP–MS) using a Nexlon2000 instrument (PerkinElmer).
Phylogenetic analysis
Protein sequences of HSP70 isoforms of different plant species were aligned using ClustalX 2.1 algorithms (Thompson et al. 2002). The phylogenetic tree was constructed using MEGA7 software with the neighbor-joining algorithm and 1000 bootstrapping replications (Kumar et al. 2016).
Heterologous expression of PtHSP70 proteins
To generate His-tagged proteins, coding sequences of PtHSP70.1 and PtHSP70.2 without stop codons were cloned into the pET28a vector and transformed into BL21. Protein expression was induced by 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 28°C for 12 h. His-tagged proteins were purified using His-tag purification resin and an affinity chromatography column (Beyotime).
Microscale thermophoresis (MST)
To investigate the Pb(II)-binding capacities of the recombinant PtHSP70-AS1 and PtHSP70-AS2 proteins, we used MST in which distinct molecules, such as protein–protein complexes versus individual proteins, respond differently to a temperature gradient (Duhr and Braun 2006; Seidel et al. 2013). The concentration of red-labeled PtHSP70-AS1 and PtHSP70-AS2 was maintained constant, whereas the concentration of the unlabeled binding partner was varied. The unlabeled binding protein was titrated at 1:1 dilutions starting at 250 µM. The samples were diluted in MST optimized buffer (pbs). The same concentrations of PtHSP70-AS1 and PtHSP70-AS2 proteins (10 µL) were again added to different concentrations of Pb(NO3)2 and fully mixed. The samples were filled into capillaries (Nano Temper Technologies), and measurements were taken after 10 min of equilibration at room temperature. The measurements were performed at auto LED power and medium MST power.
Gene identifiers
Sequence data from this article can be obtained from the Phytozome database (https://phytozome-next.jgi.doe.gov/) under the following accession numbers: PtHSP70 (Potri.010G205800), PtU1-70k (Potri.007G026900), PtUBIC (Potri.006G205700), and PtACT6 (Potri.001G309500).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession numbers PRJNA921955 (srRNA-seq) and PRJNA936161 (lrRNA-seq). The mass spectrometry proteomics data generated in this study have been submitted to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository under the data set identifier PXD039220.
Competing interest statement
The authors declare no competing interests.
Acknowledgments
This work was supported by the Natural Science Foundation of Jiangsu Province (BK20221334, SBK2020042924), the Jiangsu Agricultural Science and Technology Innovation Fund (CX (21) 2023), the Science Technology and Innovation Committee of Shenzhen (JCYJ20210324115408023), and the Major Project of Natural Science Research in Colleges of Jiangsu Province (20KJA220001).
Author contributions: Z.F.Y., C.M.X., and C.F.L. conceptualized and designed the experiments. C.X. and S.Y.C. performed experiments. C.X., S.Y.C., G.B., and C.M.X. analyzed the data. Z.F.Y. and C.X. wrote the original draft. C.F.L., C.M.X., L.P.Y.L., and Y.T. critically commented on and revised the manuscript.
Footnotes
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.277473.122.
- Received November 4, 2022.
- Accepted February 22, 2023.
This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date (see https://genome.cshlp.org/site/misc/terms.xhtml). After six months, it is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.