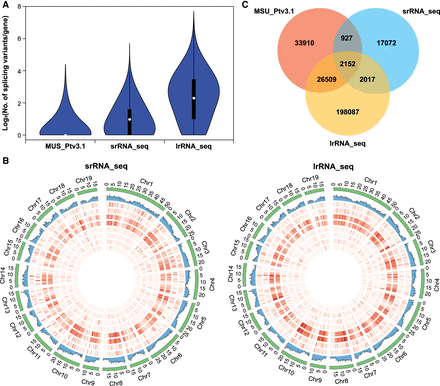
Comparison of transcript properties revealed by srRNA-seq and lrRNA-seq of Nanlin895 poplar plants (P. deltoides × P. euramericana). (A) Violin plot of spliced isoforms identified in the MSU_Ptv3.1 annotation, srRNA-seq, and lrRNA-seq data. (B) Circos representations (drawn by Circos) of post-transcriptional events identified from the srRNA-seq and lrRNA-seq data and abundance of transcripts recorded by MSU_Ptv3.1 annotation. The numbers correspond to the different layers of the Circos plot and are ordered from outwards to inwards. 1, intron retention (IR); 2, multi-IR (MIR); 3, skipped exon (SKIP); 4, multiexon SKIP (MSKIP); 5, alternative exon 5′ (AE5′); 6, alternative exon 3′ (AE3′); 7, alternative transcription start (ATS); 8, alternative polyadenylation (APA); 9, alternative 5′ first exon (AFE); 10, alternative 3′ last exon (ALE). (C) Venn diagram showing the overlapping transcripts identified from the MSU_Ptv3.1 annotation, srRNA-seq, and lrRNA-seq data.











