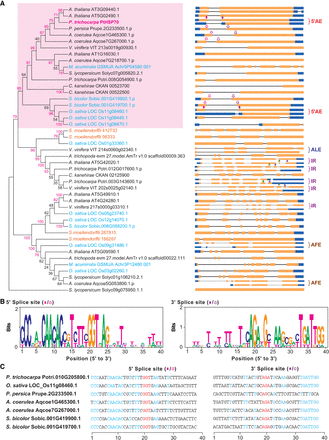
Phylogenetic and splice site analysis of HSP70. (A) Phylogenetic tree of PtHSP70 proteins. The unrooted phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replications. Blue, monocots; black, eudicots; orange, gymnosperm. The structure of each AS isoform of HSP70 genes and its corresponding splicing event are shown. Pink arrows, 5′ splice sites; purple arrows, 3′ splice sites; solid arrows, same splice sites as PtHSP70 in which AS occurs; open arrows, same splice sites as PtHSP70 but in which AS does not occur; 5′AE, 5′ alternative exon; ALE, alternative last exon; AFE, alternative first exon; IR, intron retention. (B,C) Analysis of splice site sequences of HSP70 genes. The flanking sequences of splice sites of HSP70 (Fig. 7A) were analyzed by WebLogo (B) and multiple sequence alignment (C). The conserved sequences are highlighted in red and blue.











