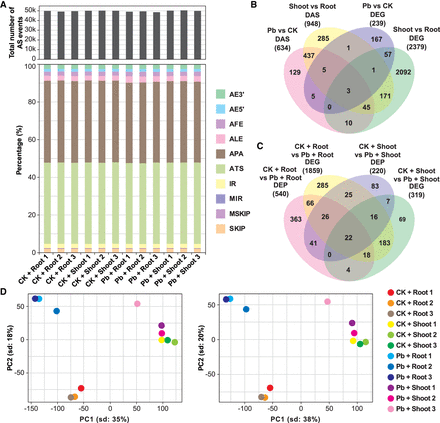
Bioinformatic comparison of the data sets of DEGs and genes subjected to DAS. (A) Summary of the total number and types of AS events identified. Data from each biological replicate are shown. (B,C) Venn diagrams displaying the common and unique genes between the data sets of DEGs and genes subjected to DAS and between the data sets of DEPs and DEGs. (D) Principal component analysis (PCA) of all 12 samples, which were divided into four groups. ALE, alternative last exon; AFE, alternative first exon; SKIP, exon skipping; MSKIP, multiple exon skipping; MIR, multiple intron retention; IR, intron retention. Group 1, CK shoots versus CK roots; Group 2, Pb roots versus CK roots; Group 3, Pb shoots versus CK shoots; Group 4, Pb shoots versus Pb roots; CK, control sample grown in the absence of Pb; Pb, sample grown in the presence of Pb.











