Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency
- Marc Friedli1,
- Priscilla Turelli1,
- Adamandia Kapopoulou1,
- Benjamin Rauwel1,
- Nathaly Castro-Díaz1,
- Helen M. Rowe1,5,
- Gabriela Ecco1,
- Carmen Unzu2,
- Evarist Planet1,
- Angelo Lombardo3,
- Bastien Mangeat4,
- Barbara E. Wildhaber2,
- Luigi Naldini3 and
- Didier Trono1
- 1School of Life Sciences and “Frontiers in Genetics” National Program, Ecole Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland;
- 2Pediatric Surgery Laboratory, Department of Pathology and Immunology, Faculty of Medicine, University of Geneva, 1211 Geneva, Switzerland;
- 3San Raffaele Telethon Institute for Gene Therapy and Vita Salute San Raffaele University, 20132 Milan, Italy;
- 4Department of Pathology and Immunology, Faculty of Medicine, University of Geneva, 1211 Geneva, Switzerland
- Corresponding author: didier.trono{at}epfl.ch
Abstract
Endogenous retroelements (EREs) account for about half of the mouse or human genome, and their potential as insertional mutagens and transcriptional perturbators is suppressed by early embryonic epigenetic silencing. Here, we asked how ERE control is maintained during the generation of induced pluripotent stem cells (iPSCs), as this procedure involves profound epigenetic remodeling. We found that all EREs tested were markedly up-regulated during the reprogramming of either mouse embryonic fibroblasts, human CD34+ cells, or human primary hepatocytes. At the iPSC stage, EREs of some classes were repressed, whereas others remained highly expressed, yielding a pattern somewhat reminiscent of that recorded in embryonic stem cells. However, variability persisted between individual iPSC clones in the control of specific ERE integrants. Both during reprogramming and in iPS cells, the up-regulation of specific EREs significantly impacted on the transcription of nearby cellular genes. While transcription triggered by specific ERE integrants at highly precise developmental stages may be an essential step toward obtaining pluripotent cells, the broad and unspecific unleashing of the repetitive genome observed here may contribute to the inefficiency of the reprogramming process and to the phenotypic heterogeneity of iPSCs.
The forced expression of a combination of transcription factors such as POU5F1 (also known as OCT4), KLF4, and SOX2 can result in the reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) (Takahashi and Yamanaka 2006; Takahashi et al. 2007). The efficiency of this process varies according to the cells chosen as starting material and the protocols used for their modification, but without further manipulation it does not exceed a few percent and it implies a latency of 1 wk to several weeks, suggesting that a cascade of events, some of which are probably stochastic, is required for full reprogramming (Jaenisch and Young 2008; Hanna et al. 2009; Yamanaka 2009). During this period, a complex sequence of still incompletely characterized epigenetic changes takes place, whereby the expression of pluripotency genes is ultimately induced whereas that of differentiation genes is repressed (Koche et al. 2011; Polo et al. 2012).
The development of a totipotent fertilized egg into a nascent embryo is a paradigm for the reverse process. It too stems from epigenetic mechanisms, which are essential not only for the establishment of specialized lineages but also for events such as imprinting and, most importantly, for the silencing of endogenous retroelements (EREs). EREs collectively account for more than half of the genome of either humans or mice, with thousands to millions of copies of endogenous retroviruses (ERVs), long interspersed elements (LINEs), short interspersed elements (SINEs; which in humans include Alu repeats), or SVAs (SINE-VNTR-Alu, a hominoid-specific ERE family) (de Koning et al. 2011). These genetic invaders, which multiply by the copy-and-paste process that defines retrotransposons, are targeted during the first few days of embryogenesis by silencing mechanisms notably involving their recognition by sequence-specific protein- or RNA-based repressors and the secondary recruitment of heterochromatin-inducing complexes (Rowe and Trono 2011). Histone methylation, histone deacetylation, and DNA methylation ensue, which inactivate their potential as insertional mutagens (Kaer and Speek 2013; Shukla et al. 2013) and repress their cis-acting transcriptional components, which would otherwise activate neighboring genes via promoter or enhancer effects (Bourque et al. 2008; Kunarso et al. 2010; Macfarlan et al. 2011; Rebollo et al. 2012; Schmidt et al. 2012; Chuong et al. 2013; Rowe et al. 2013).
The epigenetic control of EREs is a rigorously orchestrated process, with some of these elements never expressed, and others transcribed in low-cellularity embryos or in embryonic stem cells (ESCs) to be silenced only later (Lane et al. 2003; Macfarlan et al. 2011). It has recently emerged that transient transcription driven by specific coopted ERE integrants that briefly evade repression seems critical to the identity of the pluripotent state both in mice (Macfarlan et al. 2012) and humans (Santoni et al. 2012; Lu et al. 2014). This layered repression process likely reflects at least in part the orderly action of cognate repressors, including KRAB-containing zinc finger proteins (KRAB-ZFPs), which together with their cofactor TRIM28 (also known as KAP1 or TIF1B) are key to the early embryonic control of a broad spectrum of retrotransposons (Wolf and Goff 2007; Matsui et al. 2010; Rowe et al. 2010). Interestingly, KRAB-ZFPs, encoded in the hundreds by the genomes of both mouse and human, are widely expressed in ESCs but subsequently adopt highly tissue-, stage-, and cell-specific patterns of expression (Barde et al. 2013; Corsinotti et al. 2013). Whether the same holds true for other yet-to-be-identified early embryonic controllers of EREs is unknown. However, it is likely that any lag between the de-repression of specific ERE integrants and the reactivation of their sequence- or class-specific repressors will open a window of opportunity for ERE-originating transcriptional perturbations and, at least for the small fraction of these elements still endowed with retrotransposition ability (Finnegan 2012), for insertional mutagenesis. Accordingly, the present study was undertaken to examine how the transcriptional control of EREs is maintained during the reprogramming of either murine or human somatic cells, and whether it is fully reestablished in induced pluripotent stem cells.
Results
Global ERE de-repression during the reprogramming of mouse and human cells
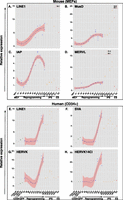
In order to assess the control of EREs during reprogramming to pluripotency, we transduced mouse embryonic fibroblasts (MEFs) harboring a Pou5f1-GFP transgene with a lentiviral vector expressing POU5F1 (OCT4), KLF4, and SOX2 (hereafter called OKS) as a single polycistronic transcript from a spleen focus forming virus (SFFV) promoter (Supplemental Fig. S1A; Pasi et al. 2011). As expected, OKS-transduced MEFs formed GFP-positive colonies after ∼12 d (Supplemental Fig. S1B) and silenced the OKS vector (Supplemental Fig. S1C). We picked and replated a series of iPSC clones and verified that they expressed Nanog at similar levels as ESCs (Supplemental Fig. S1D) and could be differentiated into embryoid bodies (EBs) (Pasi et al. 2011), indicating successful reprogramming. We then used real-time quantitative PCR (RT-QPCR) to measure the expression of families of EREs at various times of the reprogramming process and in the resulting iPSCs. We consistently observed marked increases in the transcript levels of all tested EREs in independent reprogramming experiments (Fig. 1A–D). The timing of the up-regulation was variable, usually initiating 6–10 d post-transduction with a burst of expression occasionally observed around day 2. In all iPSC clones tested, LINE1 and the ERV MusD remained highly expressed, whereas IAP (intracisternal A particle, another ERV) exhibited a fully repressed state, so that for these families of retroelements expression patterns were roughly comparable between iPSC and ESC, as previously noted (Wissing et al. 2012). This is consistent with a model whereby the trans-acting factors controlling IAPs, but not those responsible for silencing LINE1 and MusD, are reactivated during the late stages of reprogramming. MERVL, another ERV, exhibited little change in expression during reprogramming itself, but rose sharply in iPSCs, where its levels were markedly above those measured in ESCs. Rather than a defect in controlling factors, this could reflect higher fractions of cells cycling into an early post-zygotic-like state where the MERVL long terminal repeat (LTR), which serves as promoter for many genes restricted to the 2/4 cell stage of embryonic development, is particularly active (Macfarlan et al. 2012).
Global up-regulation of EREs during reprogramming. qPCR quantification of transcripts from indicated murine (A–D) and human (E–H) EREs during reprogramming of MEFs and cord blood CD34+ cells, respectively. Expression levels are indicated for parental cells (gray dots), cells transduced with an SFFV-GFP control vector (green dots), OKS-induced reprogramming time points (blue dots), individual iPS clones (orange dots), and ES cells (red dots). For the smoothing pattern across OKS-induced reprogramming time points, we computed a locally weighted polynomial regression (LOESS) with a 95% confidence interval.
We next assessed the control of EREs during the reprogramming of human somatic cells. Upon transduction of cord blood CD34+ cells with the OKS vector, iPSC clones were efficiently obtained (Supplemental Fig. S1B), which exhibited a morphology comparable to that of ESCs, expressed a full set of pluripotency genes, induced the formation of teratomas when injected into immunodeficient mice, and were karyotypically euploid (Supplemental Fig. S1E–H). However, de-repression of all tested human ERE families was detected by RT-QPCR during the reprogramming process, with sharp increases around day 19 post-transduction across three independent experiments performed with cells from different donors (Fig. 1E,H; Supplemental Fig. S2). Similar to their murine counterparts, human LINE1s remained highly expressed in pluripotent cells, as did SVAs (Fig. 1E,F). In contrast, the LTR-containing HERVK, including its HERVK14ci strain, was silenced in iPSCs, mimicking the behavior of IAPs in the murine system (Fig. 1G,H).
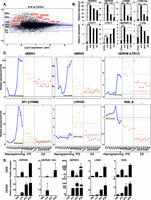
To confirm and extend these observations, we performed RNA deep sequencing (RNA-seq) at multiple time points of an independent reprogramming experiment, including in our analysis the parental CD34+ cells, six of the resulting iPSC clones, and a human ESC clone (H1) (for RNA-seq data, see Supplemental Tables 1–3, and GEO accession number GSE57866). A comparison of the transcriptomes of cells harvested at day 19 post-transduction with that of the starting population confirmed the de-repression of multiple EREs, and further identified specific subclasses of dysregulated retrotransposons (Fig. 2A), with HERVH, HERVK, and their associated LTRs LTR7Y and LTR5-Hs displaying the most pronounced up-regulation (Fig. 2B). In iPSCs, some of these elements, such as HERVK and S71 (LTR6b), displayed a repressed state comparable to that found in ESCs, but others (e.g., HERVW, LTR17, LTR7Y, L1Hs) did not or did only partially (Fig. 2B,C). Of note, while significant expression of HERVH was detected in iPSCs and ESCs, as previously reported (Santoni et al. 2012), close to a third of the HERVH integrants up-regulated at day 19 were not among those ultimately detected in these cells (data not shown). Finally, in addition to the d19 peak of expression, we occasionally observed a much earlier burst around d2–d4 for some retroelements (e.g., HERVH and HERVK) (Fig. 2C). We further RNA-sequenced eight independent human ES cell lines (UCLA1-6 [Diaz Perez et al. 2012], H19, and an independent H1 sample) to verify that the elevated ERE expression levels detected in individual iPS clones were not a general feature of pluripotent cells. We found very little heterogeneity between all tested human ES cell lines, which sharply contrasted with the inappropriate control of EREs in iPS clones and even more with their marked up-regulation during the reprogramming process (Fig. 2C). To ascertain that the observed unleashing of EREs during reprogramming is not restricted to fibroblasts or blood cells, we assessed ERE expression during reprogramming of human liver cells. We thus reprogrammed primary human hepatocytes with OKS or with a vector that also included MYC (OKSM) (Sommer et al. 2010) and measured ERE-specific transcription by real time quantitative PCR. Confirming the generality of this phenomenon, all ERE classes recapitulated the burst of expression during reprogramming previously observed in MEFs and CD34+ cells (Fig. 2D).
De-repression of individual EREs during reprogramming and in iPSCs. (A) MA-plot comparing RNA-seq–determined transcriptome of day 19 (d19) OKS-transduced vs. parental CD34+ cells. Transcripts (RefSeq) are plotted in black with the ratio (d19/CD34+) on the y-axis and expression levels on the x-axis. Representative up-regulated Repbase families are shown in red. Transversal blue lines depict magnitude of gene deregulation (e.g., only 1% of genes lie above the 99% line). (B) Expression levels of indicated HERVs in parental (average of three samples) or d19 OKS-transduced CD34+ cells, human iPS cells (average of six clones from the same reprogramming experiment), and the H1 ES cell line (average of two samples). Fold changes compared with CD34+ triplicates and P-values are calculated using the DESeq package (Anders and Huber 2010). (C) Relative expression of indicated HERVs during reprogramming of CD34+ cells, in six resulting iPSC clones (orange dots) and nine independent samples of hES cells (red dots). Green line indicates parental cells (average of three, same donor); dotted green lines, plus and minus standard deviation; and solid blue line, reprogramming time points. The horizontal (blue, orange, and red) solid lines show the mean of each group of samples. Dotted lines show a 95% confidence interval for each mean. We performed a Wilcoxon test for each mean to test if it was different from one. (D) qPCR quantification of transcripts from indicated human EREs during reprogramming of primary hepatocytes. (Top) Reprogramming with OKS. (Bottom) Reprogramming with OKSM. (Hep) Average of four nontransduced hepatocyte samples.
Deregulation of ERE-close gene transcripts during reprogramming
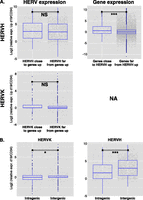
We next asked whether the up-regulation of ERVs that occurred during reprogramming impacted the expression of nearby genes. Using a twofold change cutoff and an adjusted P-value of <0.05, we identified 3703 genes up-regulated at d19 in OKS-transduced CD34+ cells compared with nontransduced cells. Genes near up-regulated HERVH (<40 kb between TSS and HERV, n = 365 genes) were far more likely (P = 2.603 × 10−05) to be up-regulated than genes more distant from these elements (>80 kb between TSS and HERV, n = 18,145 genes) (Fig. 3A). A similar analysis could not be performed for HERVK due to insufficient numbers. However, two additional lines of evidence confirmed that it was the HERV that influenced the gene and not the reverse. First, for both HERVH and HERVK, expression was not influenced by distance from an up-regulated gene (Fig. 3A). Second, intragenic HERVs were not more often up-regulated than their intergenic counterparts (Fig. 3B); in fact, the reverse trend was observed (P = 0.024 for HERVK and P = 4.26 × 10−10 for HERVH).
Up-regulation of ERE-close genes during reprogramming. (A) Using a twofold change cutoff and an adjusted P-value of <0.05, 3703 genes were found up-regulated in CD34+ cells at day 19 post-OKS transduction, compared with untransduced cells. (Left) Relative expression of HERVH (top) or HERVK (bottom) integrants situated close (<40 kb) or far (>80 kb) from these genes. No significant difference was detected (Wilcoxon test P = 0.14 for HERVK, 0.11 for HERVH). (Right) Reverse analysis, revealing that HERVH-close transcripts (n = 365 genes, or 695 transcripts) were more likely to be up-regulated than HERVH-distant transcripts (n = 18145 genes, or 30,053 transcripts). (NA) Not available due to insufficient numbers. (B) Expression levels of intragenic vs. intergenic HERVHs or HERVKs at d19 of CD34+ cells reprogramming showing no bias toward intragenic HERVs. In fact, the reverse trend was observed, with more intergenic than intragenic HERVs up-regulated at d19 of reprogramming.
Heterogeneity of human iPSC clones in repression of specific EREs and induction of prototypic ERE controllers
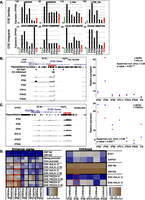
Importantly, when we compared levels of expression of individual EREs between iPSC clones derived from a single donor and issued from the same reprogramming experiment, we noticed striking differences, notably for HERVH, HERVK, and L1Hs (e.g., cf. clone 6 and clone 43 or cf. clone 2 and clone 45) (Figs. 2C, 4A). HERVK, for instance, was fivefold to ninefold more expressed in iPS2 and iPS6 compared with iPS43, iPS45, and human ES cells (Fig. 4A, top), whereas L1Hs was threefold more expressed in iPS2 than in iPS14. Interestingly, several members of the KRAB-ZFP gene family (e.g., ZNF492, ZNF649, ZNF208) exhibited marked differences in expression between iPSC clones (Supplemental Figs. S3A, S4C). A direct comparison of the transcriptomes of two iPSC clones confirmed that specific EREs and some KRAB-ZFPs were among the most discordant transcripts, suggesting that a failure to reactivate sequence-specific repressors during reprogramming might account for the lack of silencing of their target EREs in iPSCs (Supplemental Fig. S3B). Consistent with this hypothesis, Trim28 expression increased gradually during reprogramming, and Trim28 knockout MEFs failed to reprogram (data not shown). Noteworthy, known post-transcriptional controllers of retroelements (e.g., APOBEC3A and SAMHD1) (Bogerd et al. 2006; Hrecka et al. 2011; Laguette et al. 2011) were transiently induced during reprogramming (Supplemental Fig. S3A), indicating the activation of at least some genome defense mechanisms along this process.
Clonal variability of ERE control in iPSCs. (A) Comparative expression of indicated ERE families (top) in parental CD34+ cells (average of three samples), in six iPSC clones simultaneously derived from their reprogramming, and in control H1 ES cells. (Bottom) Expression levels of individual HERV integrants in same cells as A (top). (B,C) Two examples of dysregulated HERV integrants and expression level of closest genes, HHLA1 for a HERVH on chromosome 8 (B) and PRODH for a HERVK on chromosome 22 (C). (Left) Expression level tracks, as well as hES H3K9me3 and TRIM28 binding data (obtained by ChIP-seq). Note that for the PRODH locus, ChIP-seq reads could not be mapped due to the high redundancy of this region, preventing binding site calling at this location. Red arrowheads indicate HERV orientation. (Right) Expression levels of ERE–gene pairs, with Spearman correlation calculated. (D) Activation (H3K4me1 and H3K27ac) and repressive (H3K9me3) histone marks at HERVs situated close to HHLA1 and PRODH and controls (activation mark negative controls, ZNF180 and ZNF420; positive controls, POU5F1 [OCT4] and NANOG promoters; H3K9me3 negative controls, EVX1 and GAPDH; positive controls, ZNF180 and ZNF420) depicted as a heatmap from least (blue) to most (brown) enriched. Two qPCR assays (e.g., ERE PRODH 5′ and ERE PRODH 5′ #2) were designed on the 5′ and 3′ of each ERE. Note active enhancer marks on the ERE near PRODH in iPS clones 2, 6, and 14 compared with iPS clones 43 and 45, and on the ERE near HHLA1 in iPS clone 6 compared with other clones. Up-regulated ERE–gene pairs are highlighted with a red box for appropriate iPS clones.
Incomplete control of specific EREs activates the transcription of nearby genes in iPSCs
Upon scoring the expression of specific ERE integrants, we again detected considerable heterogeneity between iPSC clones (Fig. 4A, bottom). For example, a HERVH located on chromosome 8 was four- to fivefold more expressed in iPS6 compared with other iPSC clones and to human ES, while a HERVK inserted on chromosome 22 was four- to eightfold more expressed in clones 2, 6, and 14 compared with clones 43, 45, and human ESCs (Fig. 4A, bottom). EREs can epigenetically affect the local genomic landscape owing to their content in a variety of cis-acting regulatory sequences (Bourque et al. 2008; Kunarso et al. 2010; Rebollo et al. 2012; Schmidt et al. 2012; Chuong et al. 2013; Rowe et al. 2013). Correspondingly, we found numerous instances where lack of repression of a specific ERE was accompanied by up-regulation of the adjacent gene (Fig. 4B,C; Supplemental Fig. S4A,B). For example, the HHLA1 gene, situated next to the above-mentioned chromosome-8 HERVH, was significantly expressed only in iPS6 (Spearman correlation P = 0.93, P = 0.007), while expression of the PRODH gene correlated perfectly with that of the adjacent chromosome-22 HERVK among iPSC clones (Spearman correlation P = 0.96, P = 0.003) (Fig. 4B,C). Likewise, expression of KLKB1 and C9orf129 paralleled that of HERVs situated nearby (Supplemental Fig. S4A,B). Interestingly, with the exception of the HERV near PRODH (for which mapping of ChIP-seq reads was not possible), all these EREs were previously identified as bound by TRIM28 and adorned with the H3K9me3 repressive mark in human ESCs (Fig. 4B,C; Supplemental Fig. S4A,B; data not shown). Noteworthy, when differentiated to EBs or neural committed, some ERE de-repressed iPS clones regained control of specific integrants while others remained uncontrolled (Supplemental Fig. S5). For example, the ERE near PRODH in iPS6 was still locked in the highly expressed state after neural commitment even though expression of PRODH itself was reduced about threefold (Supplemental Fig. S5).
To further explore this phenomenon, we examined the state of the chromatin at several of these loci by chromatin immunoprecipitation followed by quantitative PCR (ChIP-PCR). We found histone marks typical of active enhancers (H3K4me1 and H3K27ac) near the LTRs of the HERVs situated next to HHLA1 and PRODH in iPSCs exhibiting an up-regulation of the corresponding ERE–gene pairs, but not in clones where these units were fully repressed (Fig. 4D). Conversely H3K9me3, a repressive mark that is a hallmark of KRAB/TRIM28-mediated silencing (Schultz et al. 2002), displayed the opposite pattern at these loci.
Discussion
These data demonstrate that the reprogramming of somatic cells to induced pluripotent stem cells is accompanied by a marked de-repression of endogenous retroelements from all known classes. It has recently been suggested that transcription from EREs is important to drive ES-specific transcripts, in particular MERVL in murine cells (Macfarlan et al. 2012) and HERVH in human cells (Santoni et al. 2012; Lu et al. 2014). However, while we indeed found numerous HERVH integrants up-regulated in human ES cells compared with cord blood CD34+ cells, we also noted that more than a third of the members of this group found to be activated at day 19 of reprogramming were fully repressed in ESCs or iPSCs. Thus the possible requirement of HERVH-mediated transcription during iPS reprogramming and to maintain the pluripotent state comes at a price, as control is broadly released on this family of elements. Furthermore, HERVKs, including HERVK14ci, were markedly up-regulated during reprogramming but fully repressed in pluripotent cells. For most EREs, activation reached its peak shortly before the emergence of fully reprogrammed iPSCs. Our results suggest that this phenomenon might be due at least partly to a lack of synchronization between erasure of repressive chromatin marks at EREs on the one hand and reactivation of sequence-specific trans-repressors, for instance, KRAB-ZFPs, on the other hand. Importantly, at that stage, genes situated near up-regulated EREs had a greater chance of being themselves induced, consistent with ERE-based promoter or enhancer effects (Rowe et al. 2013). It could be that this contributes to the inefficiency of reprogramming, if it results in the stochastic activation of genes affecting the path to pluripotency (Polo et al. 2012). It will be interesting to determine whether ERE activation also occurs when reprogramming efficiency is increased by depletion of the MBD3 repressor (deterministic reprogramming) (Rais et al. 2013), by coexpression of CEBPA (Di Stefano et al. 2014), or following nuclear transfer. These faster reprogramming methods may be accompanied by timely reactivation of cognate KRAB-ZFPs, which may minimize aberrant ERE reactivation.
ERE de-repression is predicted to result only rarely in mutagenic transposition events, owing both to the paucity of retroelements still endowed with transposition capacity (Quinlan et al. 2011; Finnegan 2012) and to the presence of restriction factors blocking their spread at a post-transcriptional level (Wolf and Goff 2008). However, the transcriptional perturbation of ERE-close genes may confer iPSCs or their progeny with phenotypic anomalies difficult to detect through conventional assays, such as blockade of differentiation to particular lineages, predisposition to oncogenic changes, aberrant release of bioactive molecules, or altered immunogenicity. Supporting this model, a recent comparative analysis of 49 iPSC lines derived from several human tissues detected an aberrant up-regulation of some LTR7/HERVH transcripts and neighboring genes, including HHLA1, in several differentiation-defective clones (Koyanagi-Aoi et al. 2013). Furthermore, the potential for more distal phenotypic anomalies resulting from inappropriate ERE-induced gene activation is illustrated by the recent observation that the schizophrenia-linked PRODH gene (Kempf et al. 2008) is controlled by the nearby HERVK (Suntsova et al. 2013), which we found deregulated in some iPS clones. Our findings thus warrant an in-depth survey of the genomic, transcriptional, and epigenetic state of the repetitive genome of iPSC clones, whether these are to be used for basic research or are envisioned for clinical applications.
Methods
Reprogramming
MEFs: Primary Pou5f1-GFP MEFs were prepared from E12 embryos (http://jaxmice.jax.org/strain/008214.html) and reprogrammed by transduction with OKS using four HCT116-transducing units (HC-TU) per cell as previously described (Pasi et al. 2011). CD34+ cells from human cord blood were obtained and prepared as previously described (Barde et al. 2013), before transducing 250,000 cells with OKS using 100 HC-TU per cell. After 5 d, cells were switched to mTeSR1 medium (Stemcell Technologies no. 05859) and grown on a mouse fibroblast feeder layer until reprogrammed colonies emerged (∼21 d). Individual human iPSC clones were then picked and expanded. Primary human hepatocytes were isolated from liver biopsies as previously described (Birraux et al. 2009) and plated on Matrigel or collagen before being transduced with OKS or OKSM using 20 HC-TU per cell. After 5 d, cells were switched to mTeSR1 medium (Stemcell Technologies no. 05859) and grown until reprogrammed colonies emerged (∼25 d).
qPCR
Total RNA from cells at different reprogramming time points, iPSC clones, and ES cells was extracted with TRIzol reagent (Life Technologies no. 15596-018) and PureLink micro-to-midi total RNA Purification System (Life Technologies no.12183018). cDNA was prepared with SuperScript II reverse transcriptase and real-time PCR quantification was performed with FastStart Universal SYBR Green Master (Rox; Roche no. 04913914001). Normalization was done to two or three housekeeping genes (mouse: Gapdh, Cox6a1, Tfrc; human: TFRC, B2M).
Chromatin immunoprecipitation (ChIP)-PCR
Ten million iPSCs or ESCs were immunoprecipitated as previously described (Barde et al. 2013), with TRIM28- (Abcam ab10483), H3K9me3- (Diagenode pAb-056-050), H3K27ac- (Abcam ab4729), or H3K4me1- (Diagenode pAb-037-050) specific antibodies. SYBR green qPCR was performed to quantify enrichment at specific loci.
RNA-seq
RNA-seq was performed on an Illumina HiSeq 2000 sequencer (single-read 100-cycles assay). The library was generated from 250 ng total RNA using the TruSeq RNA Sample Preparation v2 kit (Illumina). Raw reads (100-bp single-end) were mapped to the human transcriptome (RefSeq), to the human genome (hg19 assembly), and to Repbase consensus sequences using the Bowtie short-read aligner (Langmead et al. 2009) and allowing up to three mismatches, and counts were normalized to the transcript length and to the total number of reads (RPKM). Differentially expressed RefSeq transcripts and EREs were defined using the DESeq Bioconductor package (Anders and Huber 2010).
Data access
The RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE57866.
Acknowledgments
We thank S. Offner, S. Verp, and C. Raclot for technical assistance; the staff of the Lausanne University Hospital delivery room for help in obtaining cord blood; the University of Lausanne Genomics core facility for sequencing; the EPFL histology core facility, Alessandra Piersigilli, and Anne-Laure Rougemont-Pidoux for histological sections and analyses; and Vital-IT for computing. This work was financed through grants from the Swiss National Science Foundation to D.T., from the Louis Jeantet Foundation and the Novartis Consumer Health Foundation to B.E.W., from Fondazione Telethon to L.N., and from the European Research Council to D.T and L.N. (ERC 268721, KRAB’n’KAP, and ERC 249845, TARGETINGGENETHERAPY, respectively).
Footnotes
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.172809.114.
- Received January 21, 2014.
- Accepted May 23, 2014.
This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date (see http://genome.cshlp.org/site/misc/terms.xhtml). After six months, it is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.