Genomic basis of endosymbiont-conferred protection against an insect parasitoid
- 1Department of Ecology and Evolutionary Biology, Yale University, West Haven, Connecticut 06516-7388, USA;
- 2Institute of Integrative Biology, ETH Zürich, 8092 Zürich, Switzerland;
- 3EAWAG, Swiss Federal Institute of Aquatic Science and Technology, 8600 Dübendorf, Switzerland
Abstract
Bacterial endosymbionts exert a variety of beneficial effects on insect hosts. In pea aphids (Acyrthosiphon pisum), several inherited endosymbiont species protect their hosts against parasitoid wasps, which are major natural enemies. However, strains of these symbiont species vary in their ability to confer protection against parasitoids, with some conferring almost complete protection and others conferring almost none. In this study, two strains of the endosymbiont Regiella insecticola (R. insecticola 5.15 and R. insecticola LSR1) were found to differ in ability to protect pea aphids attacked by the parasitoid Aphidius ervi. Parasitism trials reveal that R. insecticola 5.15, but not R. insecticola LSR1, significantly reduced parasitoid success and increased aphid survivorship. To address the potential genetic basis of protection conferred by R. insecticola 5.15 we sequenced the genome of this symbiont strain, and then compared its gene repertoire with that of the already sequenced nonprotective strain R. insecticola LSR1. We identified striking differences in gene sets related to eukaryote pathogenicity. The protective strain R. insecticola 5.15 encoded five categories of pathogenicity factors that were missing or inactivated in R. insecticola LSR1. These included genes encoding the O-antigen biosynthetic pathway, an intact Type 1 Secretion System and its secreted RTX toxins, an intact SPI-1 Type 3 Secretion System and its effectors, hemin transport, and the two-component system PhoPQ. These five pathogenicity factors and translocation systems are hypothesized to collectively play key roles in the endosymbiont's virulence against parasitoids, resulting in aphid protection. Mechanisms through which these factors may target parasitoids are discussed.
Insects possess multiple modes of resistance against eukaryotic natural enemies (Vinson and Iwantsch 1980; Gross 1993). One insect defense against parasitoids and pathogens is the beneficial association with inherited bacterial endosymbionts (Oliver et al. 2003, 2005, 2009; Scarborough et al. 2005; Vorburger et al. 2010). A mechanistic understanding of endosymbiont-based resistance toward parasitoids is beginning to emerge, primarily due to genomic approaches (Moran et al. 2005; Degnan and Moran 2008; Degnan et al. 2009; Oliver et al. 2009). Endosymbionts known to confer resistance are currently unculturable, and thus, mutagenesis assays to identify putative pathogenicity factors are not yet possible.
The most well-understood model system in insect endosymbiont–parasitoid interactions involves the pea aphid (Acyrthosiphon pisum), its several facultative endosymbionts in the Enterobacteriaceae, and its dominant parasitoid natural enemy (Aphidius ervi). In A. pisum, some strains of the facultative endosymbiont species Serratia symbiotica and Hamiltonella defensa provide resistance against developing parasitoid larvae, independent of aphid genotype (Oliver et al. 2003, 2005). In contrast, a strain of the sister species of H. defensa, Regiella insecticola (str. 5AU), does not confer resistance to A. pisum (Oliver et al. 2003). In H. defensa, defense is dependent on a bacteriophage that codes for multiple toxin genes and lyses symbiont cells; this bacteriophage is called APSE (A. pisum secondary endosymbiont) (Moran et al. 2005; Degnan and Moran 2008; Degnan et al. 2009; Oliver et al. 2009).
In another aphid species, Myzus persicae, a strain of R. insecticola (str. 5.15), was found to cause near-complete resistance toward the parasitoid Aphidius colemani (Vorburger et al. 2010). Endosymbiont-induced resistance was confirmed by comparing naturally infected with cured M. persicae clones and by comparing uninfected with artificially infected clones in both M. persicae and also in Aphis fabae, a distantly related aphid species. Thus, resistance is associated with the presence of R. insecticola 5.15 and is not dependent on aphid genotype. However, the underlying mechanism is unknown.
In this study we address the potential genetic basis of the protection conferred by R. insecticola 5.15. A previously sequenced strain, R. insecticola LSR1 (Degnan et al. 2010), is closely related to R. insecticola 5AU, and both originated from A. pisum collected in New York, USA. Preliminary assays suggested that R. insecticola LSR1 resembled R. insecticola 5AU in failing to confer any resistance to A. ervi (NA Moran, unpubl.). To verify that R. insecticola LSR1 and R. insecticola 5.15 differ in protective ability against parasitoids, we tested both in the same host insect–parasitoid system, A. pisum and A. ervi. We sequenced the R. insecticola 5.15 genome and compared it with the genome of R. insecticola LSR1. Based on these results, we developed hypotheses for the basis of the protective phenotype observed in R. insecticola 5.15.
Results and Discussion
Parasitism trials
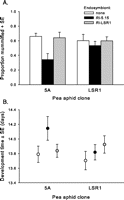
For each of two A. pisum clones, 5A and LSR1, we established three sublines: R. insecticola 5.15-infected, R. insecticola LSR1-infected, and uninfected control. Parasitism resistance assays revealed no significant difference between the two A. pisum clones, but a significant effect of subline (Table 1). This effect is attributable to a defense conferred by R. insecticola 5.15 (Fig. 1A). In the 5A aphids, infection by R. insecticola 5.15 lowered the rate of successful parasitism by about half, relative to uninfected or R. insecticola LRS1-infected lines. The effect appeared to be less in the LSR1 background; however, the clone x symbiont interaction was not significant (Table 1). Consistent with a negative effect of R. insecticola 5.15 on parasitoid development, the parasitoids that did develop to pupation in this line had a slower developmental rate as compared with those developing in R. insecticola LSR1 or uninfected lines (Fig. 1B). This was reflected in a significant subline effect in the analysis (Table 1).
Analysis of deviance (proportion mummified) and analysis of variance (wasp development time) results for the parasitism trials
Effects of R. insecticola strains on development of A. ervi in two clones of A. pisum, 5A and LSR1. (A) Proportion of parasitized wasps developing successfully in each aphid line. (B) Developmental time (days from oviposition to emergence as adults) in wasps that did develop successfully in each aphid line. Values are means +/− SE.
Genome sequencing and annotation
The 454 Life Sciences (Roche) sequencing run yielded 580,245 reads that totaled 222.91 Mbp, and assembled into 6665 contigs, corresponding to genome sequences of A. pisum, Buchnera, and R. insecticola. No lytic or lysogenic phage was present, based on both genomic sequences or PCR screening evidence. We identified 562 unique contigs comprising 2.01 Mbp assignable to R. insecticola-5.15 (Table 2). Several indicators reveal that the R. insecticola 5.15 genome sequence is near completion. First, the genome size of R. insecticola 5.15 (at 2.01 Mbp) is similar to the previously sequenced R. insecticola LSR1 (at 2.07 Mbp) (Degnan et al. 2010; Table 2). Second, the retrieved contigs included 186 of 205 single-copy genes (SICO) that are present in most Gammaproteobacteria (Lerat et al. 2003). Third, an average read coverage of 16.3× was achieved for contigs containing these genes. Assuming reads follow a Poisson distribution, >99.99% of the genome should be covered (Lander and Waterman 1988). Thus, pathogenicity factors and related metabolisms can be confidently identified for R. insecticola 5.15.
Genome comparison statistics of Regiella insecticola 5.15 with other sequenced insect-associated Enterobacteriaceae
Comparing Regiella genomes
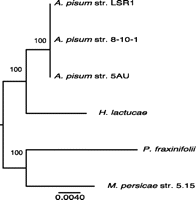
Based on phylogenetic analyses of known R. insecticola strains, R. insecticola 5.15 appears to be somewhat divergent from R. insecticola LSR1 and other strains originating from A. pisum (Fig. 2). Estimates of nonsynonymous substitutions per nonsynonymous site (dN) and synonymous substitutions per synonymous site (dS) were calculated for each SICO ortholog pair between R. insecticola LSR1 and R. insecticola 5.15 (n = 184). The mean dN is 0.0173 ± 0.05 SD, the mean dS is 0.090 ± 0.25 SD, while the mean dN/dS ratio is 0.225 ± 0.32 SD, indicating purifying selection on these core genes. These low divergence levels are consistent with classification of the strains as a single bacterial species. For comparison, mean dN and dS are much higher between sister species R. insecticola LSR1 and H. defensa (dN = 0.26, dS > 3.0) (Degnan et al. 2010).
Phylogenetic relationships of R. insecticola strains based on a 1977-bp alignment from three concatenated core genes (accD, gyrB, and murE) using RAxML with 100 bootstraps. Aphid host names are used to label branches.
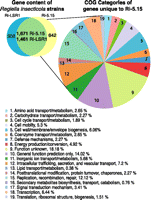
Using JGI's phylogenetic profiler and BLASTP, we identified 1671 genes in R. insecticola 5.15 that had homologs in R. insecticola LSR1, and 1461 genes in R. insecticola LSR1 that had homologs in R. insecticola 5.15 (Fig. 3). The larger number in R. insecticola 5.15 is due to two causes. First, 75.3% of multiple copies in R. insecticola 5.15 relative to R. insecticola LSR1 are due to mobile genes such as transposases, ISR elements, and repetitive hypothetical proteins, indicating greater numbers of these repetitive genes in R. insecticola 5.15. Second, 24.7% of genes with more copies in R. insecticola 5.15 involve gene fragments that are either on the edge of contigs, with truncations, or that contain premature stop codons resulting in multiple open reading frames per gene. These likely represent true pseudogenes, since such fragmentation was not found to occur in essential genes.
Comparative genomics of R. insecticola LSR1 and R. insecticola 5.15. The left pie represents the number of genes homologous and unique to each R. insecticola strain. The right pie represents the relative proportion of novel R. insecticola 5.15 genes in 19 COG categories.
We also identified 642 unique genes present in R. insecticola 5.15 with no homologs in R. insecticola LSR1 (Fig. 3; Supplemental Table 1). Of these R. insecticola 5.15-specific genes, 32.7% are hypothetical, with no significant homology with known proteins (Suppleental Table 1), whereas the remaining 67.3% had significant BLASTP hits and/or belonged to several functional gene categories (e.g., COG clusters, Pfam; see Methods) (Fig. 3; Supplemental Tables 1, 2). The large number of hypothetical proteins may partially be due to nonintact genes, since 48% of hypothetical proteins were on the edge of contigs, as expected when ORFs are interrupted by IS elements. In order to verify the status of genes unique to 5.15, we carried out PCR amplification and Sanger sequencing for apparent pseudogenes present only in 5.15 (2.2% of genes). We checked all apparent pseudogenes within contigs for cases in which two ORFs for regions of the same genes were adjacent and for cases in which a truncated gene was flanked by a hypothetical gene on the same strand (Supplemental Methods). Eight of 15 putative pseudogenes possessed errors, all in homopolymers, and these contigs were corrected.
Of the genes distinctive to R. insecticola 5.15, 39% had functional information, and these belonged to 19 COG categories (Fig. 3; Supplemental Table 2). Genes present in R. insecticola 5.15 and not R. insecticola LSR1 include unknown conserved bacterial genes, transposases, Type I, III, and IV Secretion System components, iron transporters, signal transduction systems, and phage-related proteins (Supplemental Table 2). Of the R. insecticola 5.15-specific genes, 34 encoded factors linked to eukaryote pathogenicity (Table 3). Most genes encoding pathogenicity factors were intact, with lengths of >90% of their best blastx hit homolog (Table 3). Since 454 sequencing can miscall runs of a single base, any genes with homopolymers of more than 5 nt were Sanger sequenced to confirm that errors were not present. Only one gene (prgH) possessed a homopolymer error introducing a premature stop codon, and this contig was corrected.
Gene sets of pathogenicity-related factors found in Regiella insecticola R5.15, but absent in R. insecticola LSR1
Unique pathogenicity-related factors and translocation systems
Endotoxins
The outer membrane of the Gram-negative bacterial cell wall is composed of lipopolysaccharide (LPS), consisting of Lipid A, a core oligopolysaccharide, and an O-antigen polysaccharide. LPS is referred to as an endotoxin and is associated with virulence. In particular, the O-antigen polysaccharide is important for host virulence since it is known to play roles in host cell adhesion (Jacques 1996; Jacques and Paradis 1998; Edwards et al. 2000), resistance to phagocytes (Liang-Takasaki et al. 1982), and resistance to host immune responses and surveillance (Liang-Takasaki et al. 1983; Jimenez-Lucho et al. 1987; Moran et al. 1996; Rietschel et al. 1996). Both R. insecticola strains possess similar gene sets with respect to biosynthesis of Lipid A and the core polysaccharide. In contrast, gene sets underlying biosynthesis of the O-antigen polysaccharide are drastically different between strains. R. insecticola 5.15 encodes all genes required for O-antigen polysaccharide biosynthesis, whereas the entire subpathway for dtDP-alpha-L-rhamnose (rfbABCD cluster) is missing from the genome of R. insecticola LSR1 (Table 3). As a result, the O-antigen polysaccharide is predicted to be completely absent or highly divergent in R. insecticola LSR1 (i.e., rough LPS) as compared with R. insecticola 5.15. A phylogenetic analysis of O-antigen proteins from R. insecticola 5.15 and other Proteobacteria indicates that the copy in R. insecticola may have arisen through horizontal gene transfer, as it is distantly related to the copies in other Enterobacteriaceae and more closely related to those in more distant Gammaproteobacteria and in Betaproteobacteria (Supplemental Fig. 1).
Exotoxins and Type I secretion systems (T1SS)
RTX (Repeats-in-Toxins)-related toxins are the most abundant group of exotoxins encoded in the genome of R. insecticola 5.15 (Table 4), as well as in genomes of both H. defensa 5AT and R. insecticola LSR1 (Degnan et al. 2009, 2010). RTX toxins are exotoxins secreted by the T1SS into the extracellular environment (Holland et al. 2005) that are associated with cytotoxicity and pathogenesis toward eukaryotes (Coote 1992). The genome of R. insecticola 5.15 encodes eight putative proteins with significant NCBI Conserved Domain RPS-BLAST hits to RTX toxins and related Ca2+-binding proteins (COG2931). In H. defensa and both R. insecticola strains, putative RTX (COG2931) genes also encode cysteine protease effector domains toward the N terminus for both C80 and M10 families, as in Yersinia pseudotuberculosis (Sheahan et al. 2007). R. insecticola 5.15 encodes several RTX toxins with homologs present in H. defensa or R. insecticola LSR1 (Table 4). One large putative RTX toxin (2262 amino acids, Rin_00023541) in R. insecticola 5.15 lacks homologs in either H. defensa or R. insecticola and encodes multiple serine protease domains, a Lysin domain, and COG2931 (Table 3).
Putative toxin genes found in Regiella insecticola 5.15
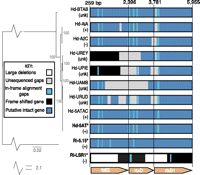
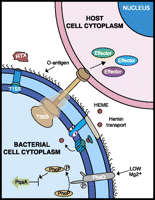
Putative RTX toxins may be functional in R. insecticola 5.15, since the genome encodes an intact T1SS operon, and TolC, an intact outer membrane protein (Rin_00000710). In contrast, R. insecticola LSR1 lacks an intact T1SS (Degnan et al. 2010). Screens of multiple H. defensa and R. insecticola strains revealed that the T1SS is intact in most H. defensa strains, but not in any R. insecticola strains, with the exception of R. insecticola 5.15 (Fig. 4) (accession nos. JF928377–JF928387). During attempts to amplify T1SS genes, only small fragments were obtained in the majority of R. insecticola strains, suggesting that genes encoding the T1SS are inactivated and fragmentary as in R. insecticola LSR1. Thus, the T1SS is uniquely present in R. insecticola 5.15 among sampled R. insecticola strains, including LSR1. The T1SS and its associated exotoxins, such as RTX toxins, are candidates for protection against parasitoids conferred by R. insecticola 5.15 (Fig. 5).
T1SS nucleotide gene alignments of H. defensa and R. insecticola strains in reference to H. defensa 5AT. Bolded strains with stars represent sequenced genomes. Phylogenetic relationships of strains are based on a 4334-bp alignment from T1SS genes (rtxB2, rtsD, rtxB1) using RAxML with 100 bootstraps. Plus signs indicate that the strain protects its aphid host against parasitoids, minus signs indicate a nonprotective phenotype, and “unk” indicates unknown phenotype, based on previous studies (Oliver et al. 2003, 2009; Vorburger et al. 2010) and this study.
Eukaryotic virulence factors present in the protective endosymbiont strain R. insecticola 5.15, but absent in the nonprotective strain R. insecticola LSR1.
Genes encoding several other exotoxins were identified in the genome of R. insecticola 5.15 (Table 4). But, in contrast to the RTX-related toxins, most of these toxins appear to be nonfunctional, and thus not important in virulence toward parasitoids. Several of these toxins are on the ends of contigs, however, and may still be intact. A gene encoding the putative hemolysin toxin (PhlA), belonging to a two-partner secretion system was identified; however, a gene encoding the secretion protein PhlB, required for exotoxin secretion into the extracellular environment (Brillard et al. 2002), was not detectable. Also, R. insecticola 5.15 encoded three truncated toxins belonging to the clostridial glycosylating toxin family and a truncated calmodulin-sensitive adenylate cyclase toxin. Another toxin encoded in R. insecticola 5.15 that is related to insect pathogenicity is makes caterpillars floppy (Mcf) (Daborn et al. 2002), for which six divergent homologs were detected, including some that are distant from homologs present in R. insecticola LSR1. One Mcf protein (Rin_00019530), located on the end of a contig, also encodes a TcdA/TcdB pore-forming Pfam domain. Another eukaryotic toxin found in R. insecticola 5.15 is a 36-amino acid putative YD-repeat toxin. This toxin is also found in R. insecticola LSR1 and appears to be truncated in both symbionts.
Type III secretion system (T3SS) and effectors
The T3SS and its associated effectors are important for many interactions between Gram-negative bacteria and eukaryotic hosts, such as attachment and invasion of host cells, the avoidance of phagocytosis, and suppression of host cell innate immunity (Hueck 1998; Waterman and Holden 2003; Matsumoto and Young 2009). As in H. defensa, R. insecticola 5.15 encodes two types of T3SS (SPI-1 and SPI-2); in contrast, R. insecticola LSR1 only encodes SPI-2 (Table 3). Several putative T3SS effectors, such as the SPI-1 effector YopJ/P, were present in R. insecticola 5.15 and not in R. insecticola LSR1 (Table 3). In Yersinia this effector is responsible for inducing apotosis and suppressing the innate immune system by targeting MAPK and NF-κB signaling pathways (Orth et al. 2000). Because the SPI-1 T3SS and associated effectors are present in R. insecticola 5.15 and not in R. insecticola LSR1, these factors are candidates for genes underlying virulence toward parasitoids (Fig. 5).
Two component systems
R. insecticola 5.15 encodes PhoPQ, a two-component signaling system that is associated with virulence in Salmonella sp., Shigella sp., Pseudomonas aeruginosa, Mycobacterium tuberculosis, Neisseria meningitidis, Y. pseudotuberculosis, and Yersinia pestis. Both R. insecticola LSR1 and the sister species, H. defensa, lack phoPQ. Phylogenetic analyses of the R. insecticola 5.15 phoPQ operon show that it is closest to homologs in other Enterobacteriaceae (Supplemental Figs. 2–4), an observation that is consistent with vertical transmission of phoPQ and loss in both H. defensa and LSR1, or with the horizontal acquisition by R. insecticola 5.15 from Yersinia or a relative.
Broadly, PhoPQ-induced phenotypes are associated with bacterial cell envelope modifications, which specifically include increased survival in macrophages, invasion of cells, tolerance to acidic pH and antimicrobial peptides, resistance to antibiotics, modification of Lipid A, and antigen presentation (Miller et al. 1989; Groisman et al. 1997; Guo et al. 1997; Ernst et al. 1999; Macfarlane et al. 1999; Moss et al. 2000; Oyston et al. 2000; Johnson et al. 2001; Groisman 2001; Perez et al. 2001; Grabenstein et al. 2004). This two-component system controls expression of the pbgPE operon in Photorhabdus luminescens and is associated with virulence against insects; pbgPE mutants are lacking an O-antigen, resulting in increased sensitivity to insect-produced antimicrobial peptides and acidic pH (Derzelle et al. 2004; Bennett and Clarke 2005).
In addition, the PhoPQ-activated, pathogenicity-related protein PqaA (Table 3) is encoded in the genome of R. insecticola 5.15, but absent from the genome of R. insecticola LSR1. Interestingly, this protein is active against the antimicrobial peptide melittin (Baker et al. 1997), found in bee and parasitoid venom (Habermann 1972; Uçkan et al. 2004). Potentially PhoPQ and PqaA play a key role in virulence toward parasitoids, possibly through PqaA-induced resistance to parasitoid venom-associated antimicrobial peptides, under PhoPQ regulation.
Iron acquisition
Availability of iron to bacteria living within host environments is generally limited; successful uptake and maintenance of iron is essential for bacterial growth and virulence (Thompson et al. 1999; Anzaldi and Skaar 2010; Watson et al. 2010; Braun and Hantke 2011). Bacterial pathogens generally possess multiple iron transport systems, depending on what type of iron is available in the host environment (Braun and Hantke 2011). In P. luminescens, iron-uptake via Yfe- and TonB-dependent transporters is required to induce virulence against insect larvae (Watson et al. 2010). The genome of R. insecticola 5.15 encodes multiple iron and hemin transport systems, such as the ABC iron transporter Yfe, the non-ABC iron transporter Feo, and the TonB-dependent hemin transporter Hmu. R. insecticola LSR1 encodes transporters Yfe and Feo, but lacks the TonB-dependent Hmu operon (Table 3). In turn, R. insecticola 5.15 encodes a diverse array of iron transporters relative to R. insecticola LSR1; potentially R. insecticola requires iron transporters to induce parasitoid virulence gene sets (Fig. 5). Alternatively, because R. insecticola 5.15 possesses a broader array of iron transporters it may be more successful than R. insecticola LSR1 in competing with the parasitoid for iron. Iron deficiency in parasitized aphids has been shown to result in arrested development of parasitoids (Zohdy 1976). Thus, bacterial competition for iron may slow wasp development, or may be lethal for the wasp.
Amino acid biosynthesis and metabolism
Larval endophagous parasitoids of aphids, such as A. ervi, feed on insect host haemolymph via ingestion and trans-cuticle absorption (Pennacchio et al. 1995; Rahbé et al. 2002; Giordana et al. 2003; Caccia et al. 2005). As in animals, generally parasitoids do not encode pathways for essential amino acid biosynthesis and must obtain these amino acids from their diet, particularly in the form of free amino acids during early larval development (Thompson 1986). In A. ervi, external sources of essential and nonessential free amino acids play a substantial role in early larval nutrition (Giordana et al. 2003; Caccia et al. 2005). Because an adequate supply of free amino acids in aphid haemolymph is required for successful A. ervi development and survival (Pennacchio et al. 1999; Rahbé et al. 2002), competition for free amino acids by R. insecticola 5.15 is a potential factor affecting parasitoid development. However, both R. insecticola strains encode similar gene sets for amino acid metabolisms and transport. Both have lost all essential amino acid biosynthetic pathways and have retained genes for corresponding active transporters. The only difference in amino acid biosynthetic abilities is that R. insecticola LSR1 and not R. insecticola 5.15 retains the pathway to synthesize threonine from aspartate. The overall similarity of amino acid metabolisms suggests that competition for limiting amino acids is not a basis of the defense conferred by R. insecticola 5.15.
Conclusions
Facultative insect endosymbionts are not essential for host survival and reproduction, but have been found to increase host fitness under particular conditions, such as attack by natural enemies (Oliver et al. 2003, 2005, 2009; Scarborough et al. 2005; Vorburger et al. 2010). Because most endosymbionts cannot be cultured axenically, genomic approaches are the primary means for elucidating the genetic mechanisms responsible for these mutualistic effects. Through comparative genomics and parasitism assays, we found a link between the symbiont-conferred protection against a wasp natural enemy and the presence of symbiont genes homologous to virulence factors known from pathogenic bacterial species. Thus, genes that underlie pathogenicity in other bacterial species are hypothesized to contribute to the observed beneficial effects of R. insecticola 5.15 on its aphid hosts, through differential effects on the wasp parasitoids.
A previous study found that R. insecticola 5.15 confers protection against the parasitoid species A. colemani in multiple aphid strains of M. persicae and in the aphid species Aphis fabae (Vorburger et al. 2010). We found that R. insecticola 5.15 also confers the aphid species A. pisum with protection against A. ervi by reducing wasp survivorship and development (Fig. 1). This ability to confer protection contrasts with previous studies on other strains of R. insecticola originating from A. pisum hosts, including R. insecticola LSR1. Collectively, these results suggest that R. insecticola 5.15 induces a generalized protective phenotype against Aphidius parasitoids in divergent aphid host species. Whether or not R. insecticola 5.15-infected aphids have lower fitness relative to uninfected aphids in an environment without parasitism pressure has yet to be tested. Aphids harboring H. defensa have lower fitness relative to uninfected aphids (Oliver et al. 2008; Vorburger and Gouskov 2011).
The mechanism of H. defensa-induced protection against A. ervi parasitoids is dependent on the association of H. defensa with the lysogenic bacteriophage APSE (Oliver et al. 2009). In this study, no lytic or lysogenic phage is present in R. insecticola 5.15, based on sequencing or PCR screens using degenerate primers for APSE sequences. Thus, the mechanisms of endosymbiont-induced resistance toward parasitoids differ between H. defensa and R. insecticola 5.15.
By comparing genomes of protective and nonprotective R. insecticola strains, we found that numerous pathogenicity-related factors and translocation systems are unique to the protective strain 5.15. In particular, R. insecticola 5.15 protection may reflect the presence of five key eukaryotic pathogencity-related factors absent from the nonprotective strain, R. insecticola LSR1 (Fig. 5). These virulence gene sets are associated with the presence of an O-antigen, an intact T1SS and its secreted RTX toxins, an intact T3SS and its effectors, hemin transport, and the two-component system PhoPQ and related genes (Fig. 5).
One unresolved question is why these candidate “eukaryotic” pathogenicity factors would selectively target and kill the parasitoid wasp and not the aphid host. One difference between the parasitoid and aphid is the mode of R. insecticola 5.15 infection. In aphids, facultative endosymbionts, such as R. insecticola 5.15, are vertically transmitted from mother to progeny during development of eggs or embryos. If R. insecticola 5.15 directly infects the wasp larva, then it must invade orally through the gut and/or through its cuticle. Different cell types and modes of bacterial infection can result in differential regulation of pathogenicity-related factors, as known for O-antigen adhesions that recognize and attach to specific eukaryotic cells (Jacques 1996), the SPI-1 T3SS that is important for invading certain host cells (Main-Hester et al. 2008), and signal-transduction systems such as PhoP-PhoQ (Groisman and Mouslim 2006). One way that bacterial pathogens can achieve virulence is by disabling, resisting, or interfering with their host's innate immune system via T3SS effectors such as YopJ and/or O-antigens. R. insecticola 5.15 may interact differentially with the wasp and aphid immune systems. For example, hymenopteran genomes, as represented by the parasitoid Nasonia vitripennis, possess different gene sets in the Gram-negative activated immune deficiency (IMD) pathway compared with aphids (Gerardo et al. 2010) (BLASTP of Nasonia in NCBI, NZ_AAZX00000000). Also, exotoxins such as RTX toxins can be very specific for a particular cell type and organism (Coote 1992).
In summary, several eukaryote-targeted pathogenicity factors are implicated as the basis for the ability of R. insecticola 5.15 to kill, or slow development of parasitoid wasps, resulting in protection of the aphid host.
Methods
Parasitism trials
In order to compare abilities of R. insecticola 5.15 and R. insecticola LSR1 to confer defense, we exposed infected and uninfected control aphid sublines to a common source of A. ervi. For each of two A. pisum clones, 5A and LSR1, we used microinjection to establish two infected sublines (Oliver et al. 2003), R. insecticola 5.15 infected and R. insecticola LSR1 infected. All infected sublines contained stable, heritable infections, and assays for parasitoid resistance were carried out at least 15 generations after infections were established. Assays were conducted to evaluate the resistance of these aphid sublines to A. ervi, using a parasitoid stock founded by wasps collected in June 2010 in Zurich, Switzerland. In each trial, we exposed 20 aphid nymphs of similar age (48–72-h-old) on a caged plant to a single female wasp for ∼6 h. We evaluated each aphid subline in 10 replicate trials, carried out in 10 randomized complete blocks. Susceptibility was quantified 10 d later as the proportion of individuals exposed to wasps that were mummified (i.e., in which wasps matured, killed the aphid, and pupated). We excluded four replicates because wasps escaped or died and failed to produce any mummies at all. We analyzed proportions with a generalized linear model (logit link function) in R 2.9.2 (R Development Core Team 2009), testing for the effects of block, aphid clone, subline, and the clone x subline interaction. Due to overdispersion, a quasibinomial error distribution with a dispersion parameter of 3.99 was used. As recommended by Crawley (2005) for quasibinomial fits, we used F-tests rather than χ2-tests for evaluating significance. Additionally, the mean development time (time from oviposition to adult emergence from mummies) of parasitoids that completed their development successfully was analyzed by ANOVA.
Genomic DNA extraction and enrichment
We extracted DNA of R. insecticola 5.15 from 10 ethanol-preserved M. persicae individuals of line 7.9 R5.15 (Vorburger et al. 2010) using Gentra Puregene Cell & Tissue Kit (Qiagen) following the manufacturer's protocol. We enriched for R. insecticola DNA by whole-genome amplification (WGA) using a modified protocol from Pan et al. (2008) and Burke and Moran (2011). The master mix, as modified from Pan et al. (2008), consisted of 0.66M of Tre D-(+)-trehalose dehydrate and 1 uL of GenomiPhi V2 enzyme mix (1 × 100 μL) (GE Healthcare UK Limited). We incubated the master mix and DNA template at 30°C for 16 h, followed by a 70°C incubation for 20 min to stop the reaction. We conducted two rounds of WGA. Genomic DNA was purified after each round of WGA with the DNeasy kit (Qiagen) following the manufacturer's protocol for “Cleanup of Genomic DNA.” To enrich for R. insecticola 5.15 DNA from total genomic DNA, we took advantage of the fact that R. insecticola 5.15 DNA is more GC-rich than either insect or primary endosymbiont (Buchnera) DNA. We used 6-mer oligonucleotide primers (eurofins, mwg | operon) with a random mixture of G and C and with two phosphorothioate-modifed nucleotides on the 3′-end for WGA. This was successful in enriching for R. insecticola 5.15 DNA, and did not appear to bias coverage toward GC-rich contigs, as the correlation coefficient between depth of coverage and % GC for contigs was low (R2 = 0.068).
Sequencing and annotation
We conducted 454 FLX pyrosequencing on the DNA from WGA. The Genomics Core facility at the University of Arizona, Tucson carried out 454 library construction and sequencing. Pyrosequencing reads were assembled with Newbler (v 2.0.0), and contigs were binned using BLAST, % GC, and read coverage as R. insecticola, Buchnera, pea aphid, mitochondrial, or contaminants (i.e., contigs with low coverage and no gammaproteobacterial BLAST hits). For annotation, we used two general methods. For the first method, R. insecticola LSR1 scaffolds were retrieved from the NCBI genbank and were used as queries in blastx searches. Unique R. insecticola contigs were used as queries in blastx searches of GenBank's NR protein database. The resulting set of putative ORFs was then annotated using evidence from similarity searches using BLASTP and Hmmr (Pfam_ls, TIGR-FAM8.0) (Bateman et al. 2004; Degnan et al. 2010). Our second method of genome annotation used the IMG-ER pipeline of the Joint Genome Institute (Markowitz et al. 2009). JGI's pipeline assigned genes to functional categories such as Pfam (Bateman et al. 2004), COG clusters (Tatusov et al. 1997), Kegg Ortholog terms (KO) (Kanehisa et al. 2004), Enzyme Nomenclature (Bairoch 2000), Gene Ontology terms (Gene Ontology Consortium 2004), InterPro (Mulder et al. 2005), and TIGRfam (Selengut et al. 2007). Annotations were largely consistent for the two methods, but the IMG-ER homology assignments were more conservative and identified more ORFs as encoding hypothetical proteins. Hypothetical genes ≥30 amino acids that showed a significant BLASTP (E-value < 1 × 10−10) hit in NCBI were reannotated based on the first method. We submitted this genome to JGI under project Id: Gi09908 and NCBI under Genome Accession: PRJNA65437; ID: 65437 using JGI's modified annotations. Homologs and unique genes of R. insecticola strains were identified using JGI's phylogenetic profiler tool using default settings.
Phage detection
We screened R. insecticola 5.15 for the presence of an APSE-like bacteriophage by conducting PCR using conserved APSE primer pairs P17/18, P23/P24, P45/51, and P35/41 (see Degnan and Moran 2008 and Supplemental Methods for PCR conditions, concentrations, and primer sequences). We also analyzed contigs from WGA for active lytic or lysogenic phage. Indicators of phage presence include large and/or high copy-number contigs encoding only phage genes.
Phylogenetic analyses and dN/dS estimates
For the R. insecticola phylogeny, we retrieved homologs of three R. insecticola core genes (accD, gyrB, and murE) from GenBank for five R. insecticola strains sampled from the aphid species A. pisum (strains LSR1, 8-10-1, and 5AU), Hyperomyzus lactucae, and Prociphilus fraxinifolii. We aligned nucleotide sequences with MacClade 4.06 OSX (Maddison and Maddison 2008). The three genes were concatenated into a single 1977-bp alignment, and the phylogeny and 100 nonparametric bootstraps were estimated using RAxML Blackbox 7.2.7 (Stamatakis et al. 2008) on the CIPRES Science Gateway V. 3.1 (Miller et al. 2009).
For the T1SS phylogeny, we retrieved homologs of R. insecticola 5.15's T1SS (genes rtxB2, rtxD, rtxB1) from the previously sequenced genomes of R. insecticola LSR1 and H. defensa 5AT. Also, we obtained T1SS gene sequences from seven H. defensa strains that are harbored within the following whitefly and aphid hosts: Bemisia tabaci (str BTAB), Uroleucon ambrosiae (str UAMB), U. reynoldense (str UREY), U. pieloui (str UPIE), A. pisum (str A2C and A1A), and Aphis craccivora (str 5ATAC). We obtained these sequences by using PCR with degenerate primers designed from H. defensa 5AT (Supplemental Methods). Using degenerate primers designed from R. insecticola 5.15, T1SS gene sequences were also obtained from R. insecticola strains from A. pisum from the United Kingdom (UK102), H. lactucae, and P. fraxinifolii. See Supplemental Methods for details on aphid hosts, primer sequences, PCR, and sequencing. Degenerate primers successfully amplified T1SS genes in all H. defensa strains. However, in some strains, not all amplicons were successfully sequenced. All partial and entire contigs of T1SS genes for R. insecticola and H. defensa strains were submitted to NCBI under accession numbers JF928377–JF928387. For all strains with complete or near complete T1SS, nucleotide sequences were aligned with MacClade 4.06 OSX (Maddison and Maddison 2008), and nongapped regions were concatenated into a single 4334-bp alignment. We conducted phylogeny estimations as above for the R. insecticola strain phylogeny.
We estimated pairwise estimates of nonsynonymous substitutions per nonsynonymous site (dN) and synonymous substitutions per synonymous site (dS) for single-copy genes that are conserved in most gammaproteobacteria (SICO) (Lerat et al. 2003). Only SICO genes >250 bp that were present in both R. insecticola 5.15 and R. insecticola LSR1 were analyzed (n = 184). We obtained R. insecticola LSR1 sequences from GenBank, aligned SICO genes with MacClade 4.06 OSX (Maddison and Maddison 2008), and calculated dN and dS estimates using the method of Goldman and Yang (1994) in PAML (Yang 1997).
Data access
Regiella insecticola 5.15's genome is available from Joint Genome Institute's public genome database, JGI-GEBA, Gold ID: Gi09908 (http://img.jgi.doe.gov/cgi-bin/geba/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=2507262005) and NCBI. (This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession AGCA01000000. The version described in this paper is the first version, AGCA01000000.) Nucleotide data from multiple R. insecticola and H. defensa strains were submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers JF928377–JF928387.
Acknowledgments
We thank Monica Tung for help with PCR and sequencing of T1SS genes, Adam Hejmowski for his help with figures, and Brian Chang for help with data entry. We also thank Charles Godfray for kindly sending us the R. insecticola strain UK102 and Jenny Herzog for help with parasitism trials. We are grateful to Rahul Raghavan and Patrick Degnan for discussions and helpful comments on the manuscript.
Footnotes
-
↵4 Corresponding author.
E-mail allison.hansen{at}yale.edu.
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.125351.111.
- Received April 28, 2011.
- Accepted September 6, 2011.
- Copyright © 2012 by Cold Spring Harbor Laboratory Press