Translation from nonautonomous type IAP retrotransposon is a critical determinant of transposition activity: Implication for retrotransposon-mediated genome evolution
Abstract
Retrotransposons constitute a major component of the genome and their proliferation significantly impacts genome evolution. Retrotransposons can propagate autonomously or nonautonomously. Nonautonomous type transposition occurs through trans-complementation by autonomous type retrotransposons. While autonomous type retrotransposons have been studied extensively, the translation products from nonautonomous type retrotransposons are not well characterized. In a previous study, we isolated both autonomous and nonautonomous type intracisternal A particle (IAP) elements from the mouse genome and established a tissue culture assay to examine trans-complementation of nonautonomous type IAP element. Using this system in the present study, we determined an active role for the translation product from nonautonomous type IAP element. Point mutations that either eliminated or truncated the IAP protein were introduced and their effects on trans-complementation were examined. Trans-complementation efficiency correlated with the expression of nonautonomous type IAP protein. The effect of nonautonomous type IAP protein was observed only when it was provided in cis, suggesting an interaction of nonautonomous type IAP protein and its transcript immediately after transcription. Interaction of autonomous and nonautonomous type IAP proteins was demonstrated by immunostaining and coimmunoprecipitation assay. Based on these findings, we propose a model in which nonautonomous type IAP protein associates with its transcript, recruits autonomous type IAP protein, and promotes the assembly of transposition competent IAP particle. The active role of the nonautonomous type IAP protein revealed in this study may provide a new insight into retrotransposon proliferation within the genome.
Retrotransposons constitute a significant proportion of the genome (∼40% in human and mouse) and their proliferation has a profound impact on genome evolution (Kazazian 2004). Recent studies demonstrated that retrotransposons are not just junk DNA but play active roles in various aspects of genome biology such as exon shuffling (Moran et al. 1999), heterochromatin formation (Lippman et al. 2004), regulation of gene expression (Whitelaw and Martin 2001), and DNA recombination (Abrusan and Krambeck 2006). Therefore, elucidating the mechanisms that regulate retrotransposon proliferation should be helpful for a better understanding of the genome.
Intracisternal A particle (IAP) element belongs to the long terminal repeat (LTR)-type mouse retrotransposon family, and ∼1000 copies exist in the genome (Kuff and Lueders 1988; Maksakova et al. 2006). Among the various types of retrotransposons throughout the genome, the IAP element provides an appropriate model system for the study of retrotransposon biology for several reasons. First, a proportion of IAP retrotransposons have retained their transposition competency, allowing the mechanisms of transposition to be investigated, which is quite difficult in most other retrotransposons that have accumulated mutations during evolution and have lost transposition competency. Second, recent reports of mutant mice deficient in genome defense systems, such as RNA interference (Kanellopoulou et al. 2005), piRNA pathway (Aravin et al. 2007), and DNA methylation (Walsh et al. 1998), showed up-regulation of IAP expression. This observation indicates that further studies of the IAP element should provide additional clues to the intricate interactions between retrotransposons and the genome.
The IAP elements are classified into either the full-length or deleted types (Kuff and Lueders 1988; Maksakova et al. 2006). The full-length type encodes gag, pro, and pol genes, whereas the deleted types harbor various internal deletions. The deleted types are further subdivided based on the location of these deletions (Kuff and Lueders 1988). Some fractions of full-length type IAP elements can transpose utilizing their own Gag, Pro, and Pol proteins, and are called autonomous. Others are called nonautonomous and can only transpose when Gag, Pro, and Pol are provided in trans from autonomous type IAP elements, which is known as trans-complementation. Transposition of the IAP element has been reported as a causative mutation for phenotypes in spontaneous mutant mice (Maksakova et al. 2006). Although 70% of IAP elements in the genome are the full-length type, most of the transposed IAP elements reported so far are the nonautonomous IΔ1 type, which has an in-frame deletion between the gag and pol regions (Kuff and Lueders 1988).
A previous study reported frequent IAP-transposition in radiation-induced mouse acute myeloid leukemia (AML) (Ishihara and Tanaka 1997). We hypothesized that IAP-transposition in AML may provide a unique experimental system to investigate the regulatory mechanisms of retrotransposon proliferation. For this purpose, we recently isolated full-length and IΔ1 type IAP elements from the AML genome and established a tissue culture assay to examine autonomous and nonautonomous type transposition (Horie et al. 2007). In the present study, we conducted mutational analysis of IAP-elements isolated in our previous study to elucidate the mechanism of transposition. We found unexpected activity in apparently defective translation product from nonautonomous type IAP elements: it functions in cis to promote transposition of nonautonomous type IAP element. The results suggest a novel mechanism of retrotransposon proliferation and provide a new aspect for the evolutionary study of genome shaping.
Results
Structure of prototype vectors for mutational analysis and scheme of transposition assay
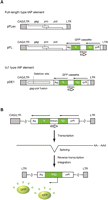
As a prototype vector for mutational analysis, we used the pFLex, pFL, and pDE1, generated in our previous study (Horie et al. 2007) (Fig. 1A, pFLex was previously called pQ14CAG). pFLex contains a full-length type IAP element isolated from radiation-induced mouse AML (Ishihara et al. 2004), and is used to express full-length type IAP protein. The pFL carries a GFP reporter cassette between the pol gene and 3′ LTR for detection of transposition. pDE1 contains a IΔ1 type IAP element isolated from the AML genome (Ishihara et al. 2004) and the GFP cassette. Sequence alignment of the pFL and pDE1 is illustrated in Supplemental Figures 1 and 2. To increase the transcription of the IAP element, the U3 region of 5′ LTR was replaced with a CAG promoter (Niwa et al. 1991). Transposition occurs autonomously in pFL; i.e., IAP protein encoded by pFL itself is sufficient for transposition and no protein from other IAP elements is required. In contrast, transposition occurs nonautonomously in pDE1 and requires trans-complementation; i.e., autonomous type IAP protein needs to be provided in trans (Horie et al. 2007). In our experimental system, autonomous type IAP protein was supplied by cotransfection of pFLex. Following introduction of point mutations into pFL and pDE1 (Fig. 2; Supplemental Figs. 1, 2), the effects of these mutations on transposition were assessed in HeLa cells. Transposition was measured by flow cytometry using GFP as a reporter, as described in our previous study (Horie et al. 2007) (see Fig. 1B for details).
Structure of the prototype vectors used for mutational analysis and experimental scheme of the transposition assay. (A) Vector structures containing the full-length type (top) and IΔ1 type (bottom) IAP element. To detect transposition, a GFP cassette is inserted in pFL and pDE1, in reverse orientation against the IAP element. The GFP gene is interrupted by an intron that is inversely oriented relative to the GFP gene. Solid and open arrows indicate orientation of the GFP gene and the intron, respectively. pFLex was used to provide full-length type autonomous IAP protein in trans to the cotransfected IAP element. The U3 region of 5′ LTR is replaced with the CAG promoter and the chimeric nature of the LTR is indicated by CAG/LTR. Pro, promoter; pA, polyadenylation signal. (B) Scheme of transposition assay. (Top) GFP is not expressed prior to transposition because of the inversely oriented intron within the GFP gene. (Middle) The intron is spliced out following transcription. GFP is not expressed from this transcript because the transcript is antisense relative to the GFP gene. (Bottom) The GFP expression unit is generated following reverse-transcription and integration into the genome. Evidence of a transposition event is detected by flow cytometry.
Amino acid changes and schematic diagram of the protein structure of mutant IAP vectors. Mutated sites and their effect on protein structure in full-length type (A) and IΔ1 type (B) IAP vectors are depicted. (A) In pFL-M1 and -M2, the first ATG and both the first and the second ATG are disrupted (indicated by ‡), respectively: pFL-M1 generates IAP protein lacking N-terminal 15 amino acids, and pFL-M2 does not generate IAP protein. Premature termination codons are introduced in pFL-M3 to pFL-M5 to truncate the IAP proteins. Gray boxes indicate predicted protein coding regions. (B) A premature termination codon is introduced in pDE1-M3 and pDE1-M11. In pDE1-M6, both the first and the second ATG are disrupted (indicated by ‡). A cluster of basic amino acids is mutated in pDE1-M12. WT, wild type. Details of mutation sites are shown in Supplemental Figures 1 and 2.
Mutational analysis of full-length type IAP element revealed unexpected activity of nonautonomous type IAP protein
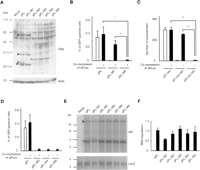
Our initial observation of nonautonomous type IAP protein activity was based on mutational analysis of a full-length type IAP element. The IAP sequence of the pFL vector was aligned with the MIA14 (or IAP14) (Mietz et al. 1987), the first isolated and most extensively studied full-length type IAP element. One of the notable differences was in a stretch of an additional 15 amino acids at the N terminus of the pFL-derived IAP protein (Supplemental Fig. 3). Since the MIA14-encoded protein appears defective in transposition activity (Dewannieux et al. 2004) and the pFL encodes an autonomously active IAP element (Horie et al. 2007), we assumed that the difference in N-terminal sequence might have been responsible for the transposition activity. To investigate the significance of the N-terminal sequence, we generated two vectors: the M1 mutant (pFL-M1), in which the first ATG start codon was eliminated, and the M2 mutant (pFL-M2), in which both the first and the second ATG codons were disrupted (Fig. 2A; Supplemental Figs. 1A, 2A). Translation from the second ATG in the M1 mutant should reduce the size of the IAP protein by only 2 kDa. Consistent with this prediction, the sizes of Gag proteins detected by Western blot were similar between pFL and M1 mutant (solid arrowheads in Fig. 3A) with only one exception (open arrowhead in Fig. 3A). In contrast, all Gag proteins detected in pFL disappeared in the M2 mutant as predicted from the presence of out-of-frame ATG triplets downstream of the mutation (Fig. 3A). Transposition activity was tested in HeLa cells using the scheme shown in Figure 1B. Autonomous transposition was nearly completely abolished not only in the M2 mutant but also in the M1 mutant (Fig. 3B), suggesting that the N-terminal sequence is critical for transposition. Transposition of the M1 mutant was successfully trans-complemented by coexpression of autonomous type IAP protein (Fig. 3B). Unexpectedly, trans-complementation was extremely inefficient in M2 mutant (Fig. 3B). To substantiate the effect of M1 and M2 mutations on transposition, we replaced the GFP reporter cassette with the neo reporter cassette used in our previous study (Horie et al. 2007). The same result was obtained with the neo reporter cassette (Fig. 3C), further confirming the effect of M1 and M2 mutations. These results suggest the importance of the nonautonomous type IAP protein, which is provided in cis from the reporter vector, in trans-complementation. The cis-effect of the nonautonomous type IAP protein was supported by the observation that the M1 mutant protein provided in trans could not improve trans-complementation of the pFL-M2 reporter vector (Supplemental Fig. 4).
Mutational analysis of full-length type IAP element. (A) Western blot analysis of Gag proteins. Arrowheads indicate Gag proteins (lanes 2 and 3). The size of the largest Gag protein in pFL and pFL-M1 is similar, probably because the N-terminal deletion of 15 amino acids in pFL-M1 reduces the size of the Gag protein by only 2 kDa, which is below the resolution limit. Alternatively, the N-terminal region of Gag is cleaved at the endoplasmic reticulum targeting signal sequence (Welker et al. 1997), which is located downstream of the deleted region. Open arrowhead (lane 2) indicates the pFL-specific band. Gag protein was not detected in pFL-M2 to pFL-M5 (lanes 4–7) except for a low molecular weight protein of unknown structure (asterisk). Mock, pBluescriptII; Actb, beta-actin. (B) Effect of mutation at the translation start site on trans-complementation examined with the GFP reporter. pFL, pFL-M1, and pFL-M2 were transfected into HeLa cells with (+) or without (–) pFLex that expresses autonomous type IAP protein, and transposition efficiency was examined by GFP signal. Data are mean ± SEM of four independent experiments. †P < 0.05 (t-test). (C) Effect of mutation at the translation start site on trans-complementation examined with a neo reporter. The GFP reporter used in (B) was replaced with a neo reporter. Transposition efficiency was examined in HeLa cells by the number of G418-resistant colonies. Transfection was conducted in triplicate and data represent mean ± SEM values. ††P < 0.01 (t-test). (D) Impaired trans-complementation by premature termination in Gag region. Data are mean ± SEM of four independent experiments. (E) Northern blot analysis of IAP transcripts from wild-type and mutated vectors. LacZ reporter was cotransfected as an internal control for transfection efficiency. Arrowhead indicates bands with predicted size, whereas asterisk denotes unpredicted bands. The absence of unpredicted bands in mock transfection (pBluescriptII and LacZ reporter) indicates that they were derived from IAP vectors. (F) Real-time RT-PCR analysis of the expression levels of IAP transcripts from wild-type and mutated vectors. Data are mean ± SEM of three independent experiments.
The cis-effect of the IAP protein was further examined with three mutant IAP reporters, each containing a termination codon at the N-terminal region of the gag gene (M3-M5; Fig. 2A; Supplemental Figs. 1A, 2A). As predicted, all Gag proteins detected in the wild-type pFL vector disappeared in all mutants (Fig. 3A). Autonomous transposition activity was completely abolished (Fig. 3D), and trans-complementation efficiency was substantially lower than M1 mutant (Fig. 3B,D). To rule out the possibility that stability of mutant IAP transcript was decreased by an RNA surveillance system such as nonsense-mediated mRNA decay (Maquat 2004), we analyzed the expression levels of IAP transcripts. Similar expression levels were observed using Northern blot analysis (Fig. 3E). Further quantitation by real-time RT-PCR showed less than twofold variation (Fig. 3F), which is significantly small compared with the difference in transposition efficiency between the vectors (Fig. 3B–D). These results further support the notion that nonautonomous type IAP protein provided in cis has an active role in transposition.
Active role of IΔ1-type IAP protein in trans-complementation
IAP-transposition has been reported in both germ cells and somatic tissues (Ishihara and Tanaka 1997; Maksakova et al. 2006). In both cases, most of the transposed elements were IΔ1 type with an internal deletion that results in in-frame fusion of gag-pol genes (Kuff and Lueders 1988). Our previous analysis of cloned IAP elements also demonstrated efficient transposition of IΔ1 type (Horie et al. 2007). Considering the significance of the cis-effect of nonautonomous type IAP protein described in the previous section, we hypothesized that a high transposition activity of the IΔ1 type may be conferred by the cis-effect of Gag-Pol fusion protein.
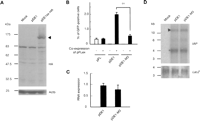
We studied the function of Gag-Pol using the pDE1 vector (Horie et al. 2007), which contains an AML-derived IΔ1 type IAP element (Fig. 1A). Since the antibody against the full-length type IAP protein did not detect Gag-Pol, we then examined the expression of Gag-Pol by inserting HA-tag at the C terminus of the Gag-Pol open reading frame. Western blot analysis confirmed the expression of HA-tagged IΔ1 type IAP protein of the expected size (Fig. 4A, arrowhead). To disrupt the expression of Gag-Pol, we generated pDE1-M3, in which a termination codon was introduced at the N-terminal region of the gag-pol gene (Fig. 2B; Supplemental Figs. 1A, 2A). As shown in the amino acid alignments of pFL and pDE1, the mutation site of the pDE1-M3 corresponds to that of pFL-M3 (Supplemental Fig. 2A). The efficiency of transposition was examined by trans-complementation assay. Similar to the mutational analysis of the full-length type vector (Fig. 3B–D), disruption of Gag-Pol expression from the reporter vector reduced transposition efficiency (Fig. 4B), suggesting a cis-effect of Gag-Pol in trans-complementation. To avoid overloading the capacity of transfected cells for accommodating transposition, we reduced the amount of DNA used for transfection. The results showed up to a sixfold difference in transposition efficiency (Supplemental Fig. 5). The cis-effect of Gag-Pol was further supported by the observation that trans-complementation efficiency did not improve when Gag-Pol was supplied in trans (Supplemental Fig. 6). We confirmed by real-time RT-PCR and Northern blot analysis that the introduction of the termination codon did not affect the IAP element at the transcription level (Fig. 4C,D).
Mutational analysis of IΔ1 type IAP element. (A) Western blot analysis of the HA-tagged IΔ1 type IAP protein. pDE1ex-HA contains HA tag at the C terminus of gag-pol open reading frame. Expected size (∼125 kDa) was observed (indicated by the arrowhead). Mock, pBluescriptII; Actb, beta-actin. (B) Effect of mutation in Gag region on trans-complementation. pFL, pDE1, and pDE1-M3 were transfected into HeLa cells with (+) or without (−) pFLex, which expresses autonomous type IAP protein, and transposition efficiency was examined by GFP signal. Data are mean ± SEM of three independent experiments. ††P < 0.01 (t-test). (C) Real-time RT-PCR analysis of the expression levels of IAP transcripts from wild-type and mutated vectors. Data are mean ± SEM of three independent experiments. (D) Northern blot analysis of the expression levels of IAP transcripts. Arrowhead indicates bands with predicted size; asterisk denotes unpredicted bands.
Although the transposition efficiency was clearly decreased by M3 mutation, pDE1-M3 retained a substantial transposition activity (Fig. 4B). This finding suggests that the nine-amino-acid N-terminal truncated Gag protein of pDE1-M3 is functional in trans-complementation. To address this point, we generated pDE1-M6 in which the first and the second ATG codons were disrupted (Fig. 2B; Supplemental Figs. 1A, 2A). No Gag-Pol expression was expected in pDE1-M6 because of the presence of multiple out-of-frame ATG triplets downstream of the mutation. The transposition efficiency of pDE1-M6 was similar to that of pDE1-M3 (Supplemental Fig. 7), suggesting that the N-terminal-truncated Gag protein of pDE1-M3 does not function in trans-complementation.
The same M3 mutation was also introduced into pDA, a IΔ1 type IAP element that was artificially generated from pFL (Horie et al. 2007). Transposition efficiency decreased sevenfold (Supplemental Fig. 8A), further confirming the cis-effect of Gag-Pol in trans-complementation. We also confirmed by real-time RT-PCR that introduction of nonsense mutation did not affect the expression level of the pDA-derived IAP transcript (Supplemental Fig. 8B).
Interaction of IΔ1-type IAP protein with full-length type IAP protein
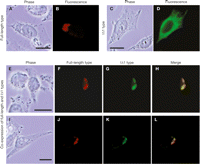
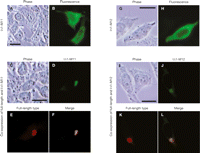
The cis-effect of the nonautonomous type IAP protein suggests that non-autonomous type IAP protein may associate with its transcript immediately after translation. Since autonomous type IAP protein is indispensable for transposition of nonautonomous type IAP element, the role of nonautonomous type IAP protein in trans-complementation could be to recruit autonomous type IAP protein adjacent to the nonautonomous type IAP transcript and facilitate the assembly of transposition competent IAP particle. If this is the case, we should observe interaction between autonomous type and nonautonomous type IAP proteins. To investigate this interaction, we ectopically expressed full-length and IΔ1 type IAP proteins in HeLa cells and analyzed spatial expression patterns by immunofluorescence. Consistent with a previous report (Welker et al. 1997), the full-length type IAP protein was observed as a perinuclear cap-like structure (Fig. 5A,B). In contrast, IΔ1 IAP protein was stained more diffusely (Fig. 5C,D). Coexpression of full-length and IΔ1 type IAP proteins diminished the diffuse pattern of IΔ1 type IAP protein, with colocalization of the two types of IAP proteins at the cap-like structure (Fig. 5E–L). The same staining pattern was also obtained in NIH3T3 cells (Supplemental Fig. 9). These results strongly suggest the interaction between full-length and IΔ1 type IAP proteins.
Subcellular localization of full-length and IΔ1 type IAP proteins. HeLa cells were transfected with the following IAP vectors: (A,B) pFLex expressing full-length type IAP protein; (C,D) pDE1ex-HA expressing IΔ1 type IAP protein; (E–L) both of pFLex and pDE1ex-HA. Major change in the localization of IΔ1 type IAP protein by coexpression of full-length type and colocalization of full-length and IΔ1 type IAP proteins suggest their interaction. Bars, 0.2 μm.
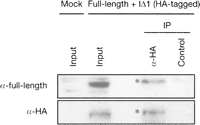
To further confirm this interaction, we coexpressed full-length and IΔ1 type IAP proteins in HEK293 cells and performed an immunoprecipitation assay. We used HEK293 cells in order to increase IAP protein expression, which increases the sensitivity of the immunoprecipitation assay. As shown in Figure 6, the full-length Gag protein was coprecipitated with HA-tagged IΔ1 type IAP protein. The result indicates interaction between full-length and IΔ1 type IAP proteins, and supports our hypothesis that nonautonomous type IAP protein mediates the assembly of IAP particle containing autonomous type IAP protein and nonautonomous type IAP-RNA.
Coimmunoprecipitation of full-length and IΔ1 type IAP proteins. HEK293 cells were transfected with pFLex expressing the full-length type IAP protein and pDE1ex-HA expressing HA-tagged IΔ1 type IAP protein. pBluescriptII was mock transfected as a control. Proteins immunoprecipitated with anti-HA antibody or with control antibody of the same subclass (anti-Oct3/4 [currently known as POU5F1]) were blotted with antiserum that detects full-length type IAP-derived Gag or with the anti-HA antibody. Migration of immunoprecipitated proteins (asterisk) is retarded compared to the input under our conditions.
Characterization of the functional region of IΔ1 type IAP protein for protein–protein interaction
To gain insight into the mechanism of trans-complementation, we tried to determine the critical region of the IΔ1 type IAP protein for interaction with the full-length type. Alignment of the full-length and IΔ1 type IAP proteins revealed significant conservation in the Pol region compared with the Gag region (Supplemental Fig. 2A,B). To determine the possible role of the Pol region in protein–protein interaction, we introduced a stop codon at the gag-pol junction of the IΔ1 type IAP protein (pDE1-M11; Fig. 2B; Supplemental Fig. 2B) and examined the interaction between the truncated IΔ1 type IAP protein and the full-length type IAP protein using the same immunostaining procedure conducted in Figure 5. The truncated IAP protein showed a diffuse expression pattern when expressed alone (Fig. 7A,B) and colocalized with the full-length type IAP protein as a perinuclear cap-like structure when expressed together (Fig. 7C–F), indicating that the Gag-region of the IΔ1 type IAP protein is responsible for the interaction with the full-length type IAP protein.
Subcellular localization of mutant IΔ1 type IAP proteins in the absence or presence of full-length type IAP protein. HeLa cells were transfected with the following IAP vectors: (A,B) pDE1ex-M11-HA expressing IΔ1 type IAP protein truncated at the Gag-Pol junction; (C–F) both pDE1ex-M11-HA and pFLex expressing full-length type IAP protein; (G,H) pDE1ex-M12-HA expressing IΔ1 type IAP protein with amino acid substitution at the basic region; (I–L) both pDE1ex-M12-HA and pFLex. Colocalization of the mutant IΔ1 type IAP proteins with the full-length type suggests their interaction. Bars, 0.2 μm.
To further narrow down the region responsible for the interaction, we searched for characteristic amino acids in the Gag region and noted a highly conserved cluster of basic amino acids (Supplemental Fig. 2A). To investigate the possible role of this region in protein–protein interaction, we replaced eight lysine residues with glutamine residues (pDE1-M12; Fig. 2B; Supplemental Fig. 2A) and examined colocalization with the full-length type IAP protein by immunostaining. The results showed that the mutation did not affect colocalization (Fig. 7G–L), indicating that the basic region is not responsible for the interaction with the full-length type IAP protein.
Although protein–protein interaction was not affected by removing the Pol region or the basic region, these mutations reduced the transposition efficiency (Supplemental Fig. 10A,B) without notable change in the stability of IAP transcripts (Supplemental Fig. 10C,D). These results suggest a possible role of these regions in other steps during trans-complementation, which will be discussed in the next section.
Discussion
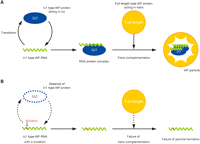
In the present study, we demonstrated that nonautonomous type IAP protein has an active role in transposition. We also showed that nonautonomous type IAP protein functions in cis to enhance transposition. Subcellular localization of IAP proteins and immunoprecipitation assay indicated interaction between autonomous and nonautonomous type proteins. Based on these results, we propose a hypothetical model in which nonautonomous type IAP protein mediates the interaction between nonautonomous type genomic RNA and autonomous type IAP protein, leading to the formation of a transposition competent IAP particle (Fig. 8).
Model for possible role of nonautonomous type IAP protein. (A) IAP particle formation mediated by IΔ1 type IAP protein. Transcript of IΔ1 type IAP element is bound by IΔ1 type IAP protein immediately after translation. The full-length type autonomous IAP protein is recruited to the RNA–protein complex through interaction between IΔ1 type and full-length type IAP proteins, leading to the formation of transposition competent IAP particle. (B) Inefficient formation of IAP particle in the absence of IΔ1 type IAP protein. Accessibility of full-length type autonomous IAP protein to IΔ1 type IAP transcript is impaired due to the lack of IΔ1 type IAP protein. Note that the conformation of the IΔ1 type IAP protein in the IAP particle is unknown and only schematically drawn here.
Dewannieux et al. (2004) reported that autonomous type IAP protein preferentially acts in cis with the IAP element that encodes it. They proposed that this cis-preference ensures preferential transposition of active IAP copies and prevents extinction of IAP elements. Our analysis of disrupted IAP protein expression in autonomous type IAP reporter (Fig. 3B–D) is consistent with their results. However, the cis-preference of autonomous type IAP protein does not account for the observation that transposition of IΔ1 type IAP elements are more frequently detected in mice than the full-length type (Maksakova et al. 2006). Identifying the active role of the nonautonomous type IAP element allowed us to design the new model shown in Figure 8, which may explain the substantial transposition activity of IΔ1 type IAP elements.
We conducted mutational analysis of the IΔ1 type IAP protein to determine the region responsible for the interaction with full-length type IAP protein. The analysis indicated that both the Pol region and the basic region in Gag are not required for the interaction (Fig. 7). On the other hand, retrotransposition efficiency was decreased in both mutants (Supplemental Fig. 10A,B), suggesting that these regions may be important in other steps during trans-complementation. One possible step would be the interaction of the IΔ1 IAP protein and genomic IAP RNA. Studies of retroviruses identified the consensus motif of Cys-X2-Cys-X4-His-X4-Cys (Covey 1986), termed the Cys-His box, in the nucleocapsid region of Gag as a potential RNA binding domain. Although two copies of Cys-His boxes exist in the full-length type IAP protein, they are absent in the IΔ1 type IAP protein (Supplemental Fig. 2A). This suggests that the previously unidentified domains could be utilized in the interaction of IΔ1 type IAP protein and genomic IAP RNA. Consistent with this concept, recent studies of HIV type I indicated that basic amino acids in the matrix region of Gag also play a role in RNA binding (Ott et al. 2005; Purohit et al. 2001). However, one should be careful when interpreting the results of mutational analysis of protein–RNA interaction because mutagenesis of IAP protein results in sequence alteration of genomic IAP RNA, which may affect the efficiency of RNA packaging into a viral particle. The mechanism of trans-complementation has been extensively studied in LINE1 (Wei et al. 2001; Garcia-Perez et al. 2007). In these studies, the authors used a series of mutant vectors in which only one or a few amino acids were mutated. A similar approach exploiting various subtle mutations will help characterize the functional domains in the IAP element.
It should be noted that factors other than the coding capacity also influence transposition efficiency. The transposition efficiency of the endogenous IΔ1 type IAP element was higher than not only the full-length type IAP element (Fig. 4B) but also the artificially generated IΔ1 type derived from the full-length type (Horie et al. 2007). This difference indicates that the high transposition activity of the endogenous IΔ1 type is due to the sequence divergence between the endogenous IΔ1 type and the full-length type (Supplemental Figs. 1, 2). The endogenous IΔ1 type IAP protein may have high activity of recruiting the full-length type IAP protein. Another possibility is that the genomic IAP RNA of the endogenous IΔ1 type is a better substrate for transposition. In support of this notion, the endogenous IΔ1 type showed substantial transposition activity upon disruption of its translation (Fig. 4B; Supplemental Fig. 7). The expression levels of IAP transcript could also be an important factor in vivo. The relative proportion of full-length and IΔ1 type IAP transcripts varies among tissues and the IΔ1 type IAP element is the major component in some tissues (Kuff and Lueders 1988), which may contribute to the high frequency of IΔ1 type IAP transposition in mice (Maksakova et al. 2006).
Some nonautonomous type retrotransposons are significantly short in size and do not appear to encode a functional protein, but are successful in propagation in the genome (Kazazian 2004). The mechanisms of such transposition have been investigated in ETn (Ribet et al. 2004), Alu (Dewannieux et al. 2003), and UnaSINE1 (Kajikawa and Okada 2002). In all cases, transposition occurs through trans-complementation by different classes of retrotransposons, for example, MusD, LINE1, and UnaL2. The mechanism of transposition proposed in the present study is different in that it requires translation of cis-acting nonautonomous type protein and the same class of autonomous type retrotransposon is utilized for trans-complementation. The next important question is to determine whether the same mechanism is also utilized in other retrotransposons in addition to the IAP element. Elucidating the active role of nonautonomous type IAP protein in transposition provides a new point of view for reevaluating the activity of retrotransposons in the genome.
Methods
Vector construction
pFL, pDE1, pDA, pFLex (previously known as pQ14CAG), and pFLneo (previously known as pQ14mneo) were described previously (Horie et al. 2007). The mutated IAP vectors depicted in Figure 2 were constructed by introducing point mutations into pFL and pDE1. To construct pDE1ex-HA and pDE1ex-M11-HA, we inserted the HA tag into pDE1 and pDE1-M11 at the C terminus of the IAP protein, and removed the GFP cassette. Details of vector constructions are described in Supplemental Methods. Sequences of primers are shown in Supplemental Table 1.
Transposition assay
HeLa cells were plated onto 12-well plates at a density of 1 × 105/well 24 h before transfection. Transfection was performed with Lipofectamine 2000 (Invitrogen) using the method recommended by the manufacturer. In each transfection, the total amount of vector DNA was adjusted to 1.6 μg with pBluescriptII (Stratagene). For flow cytometric analysis, GFP-positive cells were detected with FACScan (Becton Dickinson) 7 d after transfection and analyzed with CellQuest (Becton Dickinson) to quantitate the percentage of GFP-positive cells. Dead cells were excluded from analysis by staining with propidium iodine. For analysis based on G418 resistance, HeLa cells were maintained under nonselective medium for 5 d, followed by selection with 600 μg/mL of G418 (Invitrogen). Thirteen days later, G418-resistant cells were stained with Giemsa’s stain solution (Nacalai Tesque) and the numbers of colonies were counted. The results were presented as mean ± SEM and differences were analyzed by the Student’s t-test.
Expression and subcellular localization of full-length and IΔ1 type IAP proteins
The transfection method was similar to that used in the transposition assay. For Western blot analysis, HeLa cells were collected 2 d after transfection, suspended in sodium dodecyl sulfate (SDS) sample buffer (30 mM Tris-HCl at pH 6.8, 2.5% SDS, 10% glycerol, and 5% 2-mercaptoethanol), and cell lysates were separated on SDS-polyacrylamide gels. Proteins were transferred onto nitrocellulose membranes (Millipore) and reacted with rabbit polyclonal antiserum raised against the IAP Gag protein (a kind gift from K.K. Lueders, National Institutes of Health), anti-HA mouse monoclonal IgG (12CA5, Roche Applied Biosciences), or anti-beta-actin mouse monoclonal antibody (AC-15, Sigma), for detection of full-length type IAP protein, HA-tagged IΔ1 type IAP protein, or beta-actin, respectively. For secondary antibody, horseradish peroxidase-linked anti-rabbit or anti-mouse antibody (GE Healthcare) was used. For immunohistochemistry, cultured HeLa cells were fixed 3 d after transfection with 4% paraformaldehyde in phosphate buffered saline (PBS) for 10 min, permeabilized with 0.2% Triton X-100 (Pierce) for 10 min, and subjected to blocking with 1% BSA in PBS for 20 min. The primary antibodies used to detect full-length and HA-tagged IΔ1 type IAP proteins were the same as those used in Western blotting. For secondary antibodies, Alexa Fluor 594-conjugated goat anti-rabbit IgG (Invitrogen) and Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen) were used for full-length and HA-tagged IΔ1 type IAP proteins, respectively.
Real-time RT-PCR analysis
HeLa cells (1 × 105) were transfected with 0.6 μg of each IAP vector, and 0.2 μg of LacZ expression vector (pCMV-LacZ) as an internal control for transfection efficiency. Total RNA was purified 2 d after transfection with RNeasy Plus Mini Kit (QIAGEN), and cDNA was synthesized using random primer (Promega) and Superscript II (Invitrogen). Transcript levels from the IAP and LacZ vectors were examined by real-time PCR on a LightCycler instrument (Roche Applied Biosciences) using the LightCycler FastStart DNA Master Hybridization Probes kit (Roche Applied Biosciences). The primer pairs were 5′-GCAAGGGCAACA TCCTGT-3′ and 5′-CTGGATGAAGAAGTCGCTGA-3′ for IAP, and 5′-AAGTGACCAGCGAATACCT-3′ and 5′-GTTCAGGCAGTT CAATCAA-3′ for LacZ. The probes used were 5′-CCTTCG CCTTCGACATCCTGAGCC-fluorescein-3′ and 5′-LC Red640-GCCTTCCAGTACGGCAACCGCA-phosphate-3′ for IAP, and 5′-CTTGCCAGCGGCTTACCATCCAGC-fluorescein-3′ and 5′-LC Red640-CCACCATCCAGTGCAGGAGCTCGTTA-phosphate-3′ for LacZ. The probes for the IAP vector were designed in the GFP cassette, each residing at opposite sides of the intron sequence, so that only spliced IAP transcripts are detected. The amplification condition was 95°C for 10 min for one cycle, followed by 40 cycles of 95°C denaturation for 10 sec, 60°C annealing for 10 sec, and 72°C extension for 7 sec. The relative quantity of each transcript was obtained from the standard curve, which was drawn by serial dilutions of one of the cDNA obtained from the same set of transfection. The amount of IAP transcript was normalized by the amount of LacZ transcript.
Northern blot analysis
Northern blot analysis was performed according to the standard protocol using 8 μg of total RNA described above. A 500 bp IAP-gag fragment and a 1387 bp LacZ fragment were PCR-amplified and used as probes. The primer sequences were as follows: 5′-ATTGGTGCCGAATTCCGGGACGAGAAAATC-3′ and 5′-TCTCT CTCCAGGTATTCCTACCTAACCTTA-3′ for pFL-IAP probe, 5′-ATTGGTGCCGAATTCCGGGACGAGAAAATC-3′ and 5′-TGTA TACTTACCGTTTCTCCTTGCTCCTAC-3′ for pDE1-IAP probe, and 5′-AAATCCCGAATCTCTATCGTGCGGTGGTTG-3′ and 5′-GAAAGAAAGCCTGACTGGCGGTTAAATTGC-3′ for LacZ probe.
Immunoprecipitation assay
HEK293 cells were plated onto 60-mm dishes at a density of 1.6 × 106/dish 24 h before transfection. Cells were transfected with 4 μg of pDE1ex-HA and 4 μg of pFLex using Lipofectamine 2000 using the protocol supplied by the manufacturer. Eight micrograms of pBluescritpII was mock transfected as a control. Two days after transfection, the cells were harvested and washed three times with ice-cold PBS. After centrifugation, 7.8 × 106 cells were resuspended in 250 μL of lysis buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100) containing 1% protease inhibitor cocktail (Sigma). Cell lysates were vortexed vigorously every 10 min (repeated six times), centrifuged at 9100g for 10 min at 4°C, and supernatants were collected. They were precleared with 15 μL of protein G sepharose beads (GE Healthcare) by rotation for 30 min at 4°C. Following centrifugation at 9100g for 1 min, precleared supernatants were collected. Aliquots were stored as input, and the rest were rotated for 90 min at 4°C in the presence of 4 μg of anti-HA mouse monoclonal IgG (12CA5, Roche Applied Biosciences) or 4 μg of control monoclonal antibody of the same IgG subclass (anti-Oct3/4, C-10, Santa Cruz Biotechnology). Subsequently, 12.5 μL of protein G sepharose beads were added and rotated for 60 min at 4°C. The beads were washed four times with lysis buffer, suspended in SDS sample buffer, and analyzed by SDS-PAGE with anti-IAP serum and anti-HA mouse monoclonal IgG.
Acknowledgments
We thank K.K. Lueders for kindly providing the anti-Gag serum and H. Ishihara and J. Sakuragi for helpful discussions. E.S.S. is a JSPS Research Fellow. This work was supported in part by a grant from New Energy and Industrial Technology Development Organization of Japan and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
-
3 Present address: Department of Genetics, Cell Biology & Development/Cancer Center, University of Minnesota, Minneapolis, MN 55455, USA.
-
↵4 Corresponding author.
↵4 E-mail horie{at}mr-envi.med.osaka-u.ac.jp; fax 81-6-6879-3266.
-
[Supplemental material is available online at www.genome.org.]
-
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.069310.107.
-
- Received July 19, 2007.
- Accepted March 19, 2008.
- Copyright © 2008, Cold Spring Harbor Laboratory Press