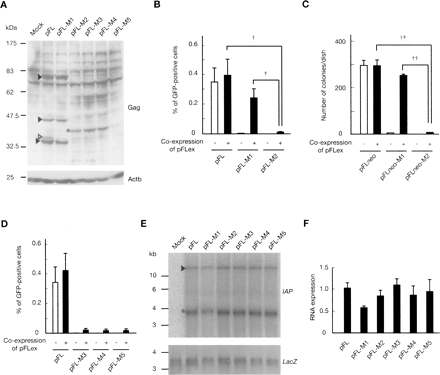
Mutational analysis of full-length type IAP element. (A) Western blot analysis of Gag proteins. Arrowheads indicate Gag proteins (lanes 2 and 3). The size of the largest Gag protein in pFL and pFL-M1 is similar, probably because the N-terminal deletion of 15 amino acids in pFL-M1 reduces the size of the Gag protein by only 2 kDa, which is below the resolution limit. Alternatively, the N-terminal region of Gag is cleaved at the endoplasmic reticulum targeting signal sequence (Welker et al. 1997), which is located downstream of the deleted region. Open arrowhead (lane 2) indicates the pFL-specific band. Gag protein was not detected in pFL-M2 to pFL-M5 (lanes 4–7) except for a low molecular weight protein of unknown structure (asterisk). Mock, pBluescriptII; Actb, beta-actin. (B) Effect of mutation at the translation start site on trans-complementation examined with the GFP reporter. pFL, pFL-M1, and pFL-M2 were transfected into HeLa cells with (+) or without (–) pFLex that expresses autonomous type IAP protein, and transposition efficiency was examined by GFP signal. Data are mean ± SEM of four independent experiments. †P < 0.05 (t-test). (C) Effect of mutation at the translation start site on trans-complementation examined with a neo reporter. The GFP reporter used in (B) was replaced with a neo reporter. Transposition efficiency was examined in HeLa cells by the number of G418-resistant colonies. Transfection was conducted in triplicate and data represent mean ± SEM values. ††P < 0.01 (t-test). (D) Impaired trans-complementation by premature termination in Gag region. Data are mean ± SEM of four independent experiments. (E) Northern blot analysis of IAP transcripts from wild-type and mutated vectors. LacZ reporter was cotransfected as an internal control for transfection efficiency. Arrowhead indicates bands with predicted size, whereas asterisk denotes unpredicted bands. The absence of unpredicted bands in mock transfection (pBluescriptII and LacZ reporter) indicates that they were derived from IAP vectors. (F) Real-time RT-PCR analysis of the expression levels of IAP transcripts from wild-type and mutated vectors. Data are mean ± SEM of three independent experiments.











