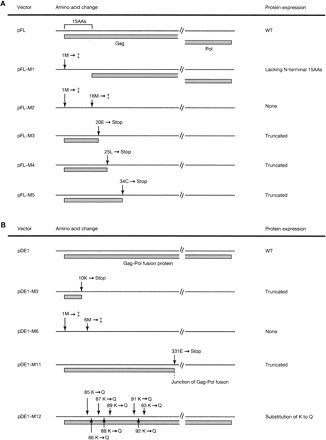
Amino acid changes and schematic diagram of the protein structure of mutant IAP vectors. Mutated sites and their effect on protein structure in full-length type (A) and IΔ1 type (B) IAP vectors are depicted. (A) In pFL-M1 and -M2, the first ATG and both the first and the second ATG are disrupted (indicated by ‡), respectively: pFL-M1 generates IAP protein lacking N-terminal 15 amino acids, and pFL-M2 does not generate IAP protein. Premature termination codons are introduced in pFL-M3 to pFL-M5 to truncate the IAP proteins. Gray boxes indicate predicted protein coding regions. (B) A premature termination codon is introduced in pDE1-M3 and pDE1-M11. In pDE1-M6, both the first and the second ATG are disrupted (indicated by ‡). A cluster of basic amino acids is mutated in pDE1-M12. WT, wild type. Details of mutation sites are shown in Supplemental Figures 1 and 2.











