The function and expansion of the Patched- and Hedgehog-related homologs in C. elegans
Abstract
The Hedgehog (Hh) signaling pathway promotes pattern formation and cell proliferation in Drosophila and vertebrates. Hh is a ligand that binds and represses the Patched (Ptc) receptor and thereby releases the latent activity of the multipass membrane protein Smoothened (Smo), which is essential for transducing the Hh signal. In Caenorhabditis elegans, the Hh signaling pathway has undergone considerable divergence. Surprisingly, obvious Smo and Hh homologs are absent whereas PTC, PTC-related (PTR), and a large family of nematode Hh-related (Hh-r) proteins are present. We find that the number of PTC-related and Hh-r proteins has expanded in C. elegans, and that this expansion occurred early in Nematoda. Moreover, the function of these proteins appears to be conserved in Caenorhabditis briggsae. Given our present understanding of the Hh signaling pathway, the absence of Hh and Smo raises many questions about the evolution and the function of the PTC, PTR, and Hh-r proteins in C. elegans. To gain insights into their roles, we performed a global survey of the phenotypes produced by RNA-mediated interference (RNAi). Our study reveals that these genes do not require Smo for activity and that they function in multiple aspects of C. elegans development, including molting, cytokinesis, growth, and pattern formation. Moreover, a subset of the PTC, PTR, and Hh-r proteins have the same RNAi phenotypes, indicating that they have the potential to participate in the same processes.
The membrane protein Patched (Ptc) and its ligand, the morphogen Hedgehog (Hh), control numerous processes during embryonic development, ranging from segmentation in Drosophila to neural tube differentiation and axon guidance in vertebrates (Ingham and McMahon 2001; Wetmore 2003). Hh is also involved in regulating stem cell maintenance and proliferation (Taipale and Beachy 2001; Zhang and Kalderon 2001). In humans, dysfunction of Hh/Ptc signaling is associated with a variety of malignancies. Sporadic PTCH mutations can cause basal cell carcinomas and medulloblastomas (Johnson et al. 1996), and hereditary mutations are responsible for Gorlin's syndrome, which is associated with developmental abnormalities and a pre-disposition to develop cancer (Hahn et al. 1996).
Studies to date indicate that signal transduction mediated through Hh and Ptc requires Smoothened (Smo), a seven-pass membrane protein related to the Frizzled family of Wingless receptors (Ingham and McMahon 2001; Lum and Beachy 2004). In the absence of Hh, Ptc represses the intrinsic signaling activity of Smo. It has been proposed that Ptc regulates the transport or the distribution of an endogenous small molecule capable of influencing Smo activity (Chen et al. 2002; Taipale et al. 2002), although earlier studies indicated that Ptc repressed Smo activity via a direct protein–protein interaction (Stone et al. 1996; Murone et al. 1999). Inhibition of Smo is released when Hedgehog (Hh) binds to Ptc. In turn, the Hh signal is transduced and Cubitus interruptus transcriptionally activates Hh targets.
The genome of Caenorhabditis elegans encodes two PTC homologs, PTC-1 and PTC-3, and a pseudogene, PTC-2 (Kuwabara et al. 2000). In addition, 24 proteins with sequence and topological similarities to PTC have also been identified and named PTR (for Patched-related). The PTC and PTR proteins are distinguished by the presence of sterol sensing-domains (SSDs) (Kuwabara et al. 2000). A common feature shared by many SSD proteins is their participation in processes that variously involve the transport of lipids, sterols, or sterol-modified proteins (Kuwabara and Labouesse 2002). Despite the presence of PTC homologs, database searches of the complete C. elegans genome sequence have failed to identify obvious Hh or Smo homologs (The C. elegans Sequencing Consortium 1998), so the role of the PTC and PTR proteins in C. elegans is unclear. However, we previously demonstrated that PTC-1 is required in the C. elegans germline for cytokinesis and maintenance of autonomous germ cell development within the gonad syncytium (Kuwabara et al. 2000).
In Drosophila and vertebrates, Hh is a bipartite protein that follows a three-step maturation process to achieve full biological activity (Jeong and McMahon 2002). The C-terminal “Hog” domain autoproteolytically cleaves the Hh precursor to generate a secreted N-terminal “Hedge” signaling activity, which is modified at the C terminus by the addition of cholesterol and at the N terminus by palmitoylation (Pepinsky et al. 1998; Chamoun et al. 2001). The genome of C. elegans encodes >60 proteins that will be collectively referred to as Hedgehog-related (Hh-r) (Bürglin 1996; Aspöck et al. 1999), although they bear no direct sequence similarity with the “Hedge” of Hh (Aspöck et al. 1999). It has been proposed that the C. elegans Hh-r proteins are the evolutionary homologs of Hh (Aspöck et al. 1999), because some of the C. elegans Hh-r proteins are also bipartite, carrying a C-terminal Hog domain and an N-terminal “Hedge-related” sequence that could have signaling potential (Aspöck et al. 1999). In addition, C. elegans encodes a number of proteins consisting only of the secreted Hedge-related domain and a single protein with only a Hog domain. The Hedge-related domains have been further divided into families based on sequence: warthog (wrt), groundhog (grd), ground-like (grl), and quahog (qua) (Bürglin 1996; Aspöck et al. 1999).
Given the importance of Hh signaling in other organisms, the absence of obvious Hh and Smo homologs in C. elegans has raised numerous questions about the roles played by the C. elegans PTC, PTR, and Hh-r proteins in development. To gain insights into the functions of these genes, we surveyed the phenotypes produced by RNA-mediated interference (RNAi) of the ptc, all 24 ptr, and 27 hh-r genes, and we show that these genes are functional and participate in multiple aspects of C. elegans development, despite the absence of Smo. We find that some of the ptc, ptr, and hh-r genes participate in the same processes, which share in common the need to transport proteins, lipids, or sterols. Although most of the ptr genes are individually capable of developing an RNAi phenotype; we demonstrate that some members share partial functional redundancy. We show that the expansion of the ptr family appears to be a feature of Nematoda as Drosophila, mouse, and human each have only a single ptr gene. Moreover, the activities of the ptc and ptr genes are conserved between C. elegans and Caenorhabditis briggsae. Finally, we provide evidence that some of the ptr genes participate in processes requiring sterols and the trafficking of proteins, sterols, and lipids.
Results
The ptc and ptr genes promote molting
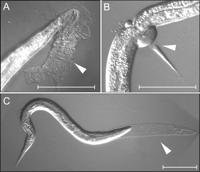
The phenotypes produced by inhibiting the activities of the ptc-1, ptc-3, and the 24 ptr genes were surveyed by RNAi using the wild-type N2 Bristol and rrf-3, a mutant hypersensitive to RNAi (Simmer et al. 2002, 2003). Table 1 reveals that RNAi inhibition of many of these genes produces a Mlt (molting-defective) phenotype. Mlt animals have problems in shedding old cuticle, which is the collagenous outer layer of the hypodermis (Fig. 1). Thus, animals retaining remnants of old cuticle, displaying circumferential constrictions, or dying from starvation because they are encapsulated in old cuticle are scored as Mlt.
The C. elegans ptc and ptr genes promote molting
When RNAi was repeated by using rrf-3, the number of ptr genes generating a Mlt phenotype more than doubled; we found that ptc-1, ptc-3, and 17 ptr genes all produced a Mlt phenotype (Table 1). For most genes, the use of rrf-3 also increased the penetrance of the Mlt phenotype and made it possible to detect Mlt worms earlier in development (Table 1). However, rrf-3 mutants are not Mlt (Table 1). It is interesting to note that the absence of lrp-1/megalin, which encodes a receptor required for cholesterol endocytosis and is also capable of endocytosing Sonic Hh, also produces a similar RNAi Mlt phenotype (Yochem et al. 1999; McCarthy et al. 2002).
Redundancy in the activity of ptr genes
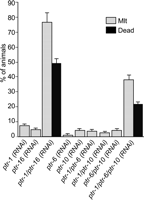
Large gene families can contain members that are functionally redundant (Prince and Pickett 2002). To determine whether this is true of the ptr family, we selected six genes—ptr-1, -6, -10, -16, -20, and -21—that individually develop a weak RNAi Mlt phenotype in rrf-3 (Table 1), and we systematically analyzed the effect of simultaneously disrupting combinations of two or three of these genes by RNAi. Of the 35 possible combinations, the simultaneous RNAi disruption (combi-RNAi) of ptr-1:ptr-16 or ptr-1:ptr-6:ptr-10 had a striking synergistic effect; the severity and the percentage of animals displaying a Mlt phenotype was enhanced (Fig. 2). Thus, some members of the ptr gene family are partially redundant in function.
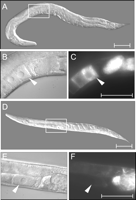
Molting defects (Mlt) associated with ptr(RNAi). (A–C) Examples of Mlt defects (arrowheads) associated with ptr-5, ptr-4, and ptr-21(RNAi), respectively. Scale bar, 50 μm.
The ptr genes are partially redundant. All 35 double and triple combinations of ptr-1, -6, -10, -16, -20, and -21 were subjected to RNAi in rrf-3; however, only the single disruptions and those combinations showing synergistic phenotypic enhancement are shown. Gray bar indicates Mlt animals; black bar, dead animals.
The ptr genes promote growth and cell patterning in hermaphrodites and males
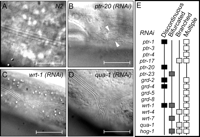
The ptr genes not only are involved in molting but also are active in C. elegans growth and morphogenesis (Fig. 3). RNAi disruption of 15 ptr genes impairs cell growth and produces Sma (small) animals that are up to 40% shorter than same-stage N2 animals (Fig. 3A,E). Inhibition of four ptr genes also leads to vulval defects (Fig. 3B,E).
The genome-wide RNAi screens, performed so far in C. elegans, have focused on elucidating the RNAi phenotypes produced by hermaphrodites and not by males. To explore whether members of the ptr family have male-specific functions, we inhibited all of the genes listed in Table 1 by RNAi using the male producing strains, him-8 or him-8;rrf-3. Inhibition of ptr-1, -2, -4, -18, -20, and -23 by RNAi produced male molting defects (Supplement 1). Disruption of these genes also caused defects in the patterning of the spicules, fan, and sensory rays of the male tail; in the most severe cases, these structures were absent (Supplement 1; data not shown). Thus, at least six ptr genes promote male tail patterning.
ptr-2 is involved in somatic cytokinesis
ptr-2(RNAi) produces a phenotype that is markedly distinct from that of all other ptr genes. Disruption of ptr-2 by RNAi causes embryos to arrest with multinucleated cells arising from a probable defect in somatic cytokinesis (data not shown). However, animals that escape from embryonic lethality (Emb) are also Mlt (Table 1). Similarly, ptc-1(RNAi) embryos arrest from defects occurring during germline cytokinesis (Kuwabara et al. 2000); however, when ptc-1(RNAi) is performed on post-embryonic animals, animals can also display molting defects (Table 1). It should be noted that ptc-1(RNAi) animals do not display defects in somatic cytokinesis.
Disruption of ptr gene activity impairs exocytosis and adult alae development
A re-examination by Nomarski DIC revealed that RNAi inhibition of 18 ptr genes caused the accumulation of abnormal fluid-filled vacuoles in the intestine and hypodermis (Fig. 3C,E). These animals were also Unc because of their tendency to curl and to become immobile as adults, although they remained sensitive to touch. This immobile Unc phenotype strongly resembles that displayed by sec-23 (yeast SEC homolog) and lrp-1 mutants, which are known to be deficient in vesicle trafficking (Yochem et al. 1999; Roberts et al. 2003). A second, apparently unrelated Unc phenotype was also detected, whereby animals generate sinusoidal body waves but fail to progress forward or backward; this defect is probably caused by tears in the cuticle (data not shown). In addition, disruption of three ptr genes led to the formation of cuticular blisters (Bli) that might result from defects in the structure of the cuticle or the failure to secrete proteases that process collagen precursors (Fig. 3D,E; Thacker et al. 1995).
CHE-14, the C. elegans homolog of the SSD protein Disp, promotes the exocytosis of proteins from the hypodermis; animals lacking che-14 display severe defects in the formation of alae (Michaux et al. 2000). The adult alae are secreted by seam cells and are composed of three raised cuticular ridges that form lateral stripes above the seam cell cords. Given the potential involvement of many ptr in trafficking, we asked whether the ptr genes could affect alae formation. We found that disruption of six ptr genes by RNAi led to the generation of four classes of defective alae: (1) multiple alae composed of four instead of three cuticular ridges, (2) discontinuous alae, (3) bifurcated alae, and (4) multiply branched alae (Fig. 4). We propose that similar to che-14, the defects in alae development resulting from ptr(RNAi) might be caused by an impairment in protein secretion (Michaux et al. 2000). Hence, it is possible that the subset of ptr genes affecting alae formation could have properties that are more similar to that of che-14/Disp than to ptc.
Disruption of ptc and ptr genes by RNAi leads to growth and developmental defects. (A)A ptr-4(RNAi) adult hermaphrodite (bottom) is 30% shorter than is a similarly staged wild-type hermaphrodite (top). (B) An everted vulva in a ptr-11(RNAi) animal. (C) ptr-1(RNAi) animals accumulate fluid-filled vacuoles in the hypodermis. (D) Blisters caused by ptr-3(RNAi). (E) Summary of phenotypes produced by RNAi, not including molting. All the ptc and ptr genes were examined; however, only those showing RNAi phenotypes are displayed. Scale bar, 50 μm.
Elimination of ptr or hh-r gene activity by RNAi causes defects in adult alae formation. (A) Wild-type alae in N2. (B–D) Defective alae produced by RNAi of ptr or hh-r gene members. (B) Discontinuous alae (arrowheads). (C) Bifurcated alae. (D) Multiply branched alae. (C, D) Multiple alae composed of four instead of three cuticle ridges. (E) Summary of alae defects after RNAi of ptr or hh-r genes. All genes displayed in Tables 1 and 2 were examined; however, only those showing phenotypes are displayed. Scale bar, 10 μm.
Inhibition of ptr-4, ptr-18, and ptr-23 disrupts endocytosis
To explore the potential involvement of the ptr genes in trafficking, we asked whether disruption of the ptr genes inhibits endocytosis. To address this question, we used an assay developed by Grant and Hirsh (1999) that examines the ability of oocytes to endocytose a fluorescent yolk protein reporter, YP170::GFP. Normally, the yolk protein vitellogenin is secreted by the intestine and endocytosed by oocytes proximal to the spermatheca (Grant and Hirsh 1999). We found that the ability of oocytes to endocytose the YP170::GFP fusion protein is compromised in adult ptr-4(RNAi) animals; instead, ectopic vacuoles of yolk collect in the body cavity (Fig. 5). A milder form of this defect is present in ptr-18(RNAi) and ptr-23(RNAi) animals, which form ectopic vacuoles of yolk (data not shown).
The hh-related ptc, and ptr genes have similar RNAi phenotypes
In Drosophila and vertebrates, Ptc interacts directly with Hh to control cell patterning and proliferation (Incardona et al. 2000). To investigate whether the C. elegans Hh-r proteins have the potential to participate in similar activities as the PTC and PTR proteins, we performed a global survey of the RNAi phenotypes produced by a subset of 27 hh-r genes (Table 2). We observed that the disruption of these genes by RNAi produces phenotypes very similar to that seen with ptc-3(RNAi) and ptr(RNAi) (Table 2; Fig. 4), including defects in molting, growth, and vulval and male tail morphogenesis (Table 2, Supplement 2). In a few cases involving individual members of three paralogous gene pairs—grd-3/grd-4, grd-5/grd-10, and grd-13/grd-14—that share >70% identity at the nucleotide level, RNAi phenotypes are likely to arise from reducing the activity of both paralogs.
RNAi phenotypes produced by a subset of hh-r genes
Expression patterns of the ptc, ptr, and hh-r genes
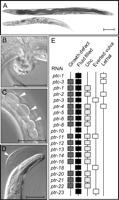
Expression profile analysis performed on selected hh-r genes revealed that they are expressed in a variety of tissues, including the hypodermis, seam cells, neurons, and neuron-associated cells (Aspöck et al. 1999). Because the ptc, ptr and hh-r genes share similar RNAi phenotypes, we sought to determine if they were expressed in the same tissues by examining serial analysis of gene expression (SAGE) data obtained from the Genome BC C. elegans Gene Expression Consortium (http://elegans.bcgsc.bc.ca/perl/sage). Although there was some diversity in their expression profiles, most members of the ptc, ptr, and hh-r genes are expressed in the same cells (Supplement 3). In particular, the transcripts of ptc-3, eight ptr, and 10 of 27 hh-r genes under examination are enriched in hypodermal cells, the site where cuticle components are synthesized and secreted between molts (Supplement 3) (Johnstone 2000). We were unable to find additional expression data in the C. elegans fusion library database (http://nematode.bsgsc.ca) or the Nematode Expression Pattern Database (NEXTDB) (http://nematode.lab.nig.ac.jp). Taken together, the similarities between the RNAi phenotypes and the SAGE profiles allow for the possibility that the PTC, PTR, and Hh-r proteins act in the same pathways.
Expansion of the ptr gene family in C. elegans
The SSD family of proteins control multiple aspects of cholesterol biosynthesis and homeostasis and also participate in processes involving the transport of proteins and lipids (Kuwabara and Labouesse 2002). We previously showed that C. elegans encodes a seventh family of SSD proteins, consisting of the 24 ptr genes (Kuwabara et al. 2000). Given that complete genome sequences for Drosophila, mouse, and human were not available at the time of our initial analysis (Adams et al. 2000; Lander et al. 2001; Waterston et al. 2002), we performed BLAST analysis to identify all SSD-containing proteins encoded by these genomes, and aligned the sequences obtained by using Clustal W (Fig. 6) (see Experimental Procedures). Similar to the results obtained for C. elegans, we found that the SSD proteins cluster into seven families, although the genomes of fly, mouse, and human each encode only a single PTR homolog. These results indicate that the ptr genes are conserved across phyla, although the family has expanded in C. elegans (Fig. 6).
ptr-4(RNAi) disrupts endocytosis. (A–C) A YP170::GFP reporter is expressed in the intestine and endocytosed by the proximal-most oocyte (arrowheads) in wild-type hermaphrodites, as visualized by GFP fluorescence in C. (D–F) ptr-4(RNAi) hermaphrodites inefficiently localize YP170::GFP to oocytes (arrowheads). Panels A, B, D, and E are Nomarski DIC micrographs; panels C, F show YP170::GFP fluorescence. Scale bar, 50 μm.
ptr gene expansion is a feature of Nematoda
The finding that the ptr genes are active during multiple aspects of C. elegans development and that their functions are not strictly redundant led us to ask whether they are present in other nematodes. We first assessed the orthology between the SSD proteins of C. elegans and C. briggsae, a rhabditid with a complete genome sequence that diverged from a common ancestor of C. elegans between 60 and 100 millions years ago (Stein et al. 2003). This was achieved by assigning each full-length C. elegans SSD protein a C. briggsae ortholog partner by reciprocal best BLAST (Stein et al. 2003). Second, we compared the syntenic conservation of genes flanking either side of each SSD encoding gene in C. elegans and C. briggsae (Kuwabara and Shah 1994). According to these criteria, all 31 C. elegans SSD proteins have a direct C. briggsae ortholog (Table 3; data not shown). Moreover, 23 of the 31 orthologous proteins share >80% amino acid identity (Table 3; Stein et al. 2003).
Orthologous pairing of C. elegans and C. briggsae SSD proteins
To ask whether PTC and PTR proteins are conserved in nonrhabditid branches of Nematoda, we searched the Brugia malayi genome sequence repository (http://www.tigr.org/tdb/parasites/); B. malayi is a filarial parasite and a member of the Spirurida order (Blaxter et al. 1998). Reciprocal best BLAST searches using predicted full-length C. elegans PTC and PTR sequences show that B. malayi encodes proteins with strong homology to C. elegans PTC-1, PTC-3, and at least 16 distinct PTR proteins (Supplement 4). We also searched for sequences encoding PTC and PTR proteins from the 3334 ESTs available for the nematode Meloidogyne arenaria, a plant parasite and member of the Tylenchida order (http://www.nematode.net) (Blaxter et al. 1998). Reciprocal best BLAST analysis led to the identification of one PTC protein and at least two M. arenaria proteins sharing strong homology to PTR-2 and PTR-4 (data not shown). Thus, the presence of multiple ptr genes in nonrhabditid clades indicates that the expansion of ptr genes is a feature of Nematoda. However, assignments of orthology remain tentative because there is insufficient sequence data available to analyze syntenic associations, except in the case of C. briggsae.
Conservation of function between C. elegans and C. briggsae ptc and ptr genes
To provide evidence that the functions of the ptc and the ptr genes are conserved between C. elegans and C. briggsae, we disrupted the activities of the C. briggsae orthologs of ptc-1, ptr-4, ptr-18, and ptr-23 by RNAi. Although the RNAi response in C. briggsae appeared to be less robust than in C. elegans, similar RNAi phenotypes were observed. Disruption of the C. briggsae ptc-1 ortholog led to the appearance of multinucleate germ cells arising from a probable defect in germline cytokinesis, and molting defects were observed in Cb-ptr-4(RNAi), Cb-ptr-18(RNAi), and Cb-ptr-23(RNAi) animals (Kuwabara et al. 2000) (Supplement 5; data not shown). These results indicate that gene activity appears to be conserved between orthologous gene pairs.
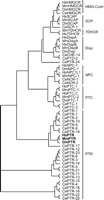
Sterol sensing domain (SSD) proteins and their phylogeny. Full-length SSD proteins can be clustered into seven major families by Clustal W: HMG-CoAr, SCP, 7-DHCR, Disp, NPC, PTC, and PTR (Thompson et al. 1997, Kuwabara et al. 2000). Sequence prefix denotes species: C. elegans (Ce), D. melanogaster (Dm), M. musculus (Mm), and H. sapiens (Hs). GenBank accession numbers are available in the Methods section.
Discussion
Homologs of the Hh signaling pathway in C. elegans
The Hh pathway has undergone extensive divergence in C. elegans, so we performed an RNAi survey to understand how the >60 Hh-r, 24 PTC-related proteins, and two PTC proteins function in the absence of Smo and Hh. Because the PTC and PTR proteins share overlapping functions, they will be discussed as a unit, although Clustal W analysis indicates that they can be subdivided into distinct SSD classes. In particular, ptc-1 and ptr-2 promote cytokinesis, and the ptc-1, -3, and most ptr genes are involved in molting (Kuwabara et al. 2000).
Our global survey of the RNAi phenotypes of the C. elegans ptc, ptr, and hh-r genes reveals for the first time that they share in common an involvement in cell growth, patterning, and molting, a process that is dependent on the availability of sterols. Molting plays an essential role in C. elegans growth and proliferation. This process involves the formation and degradation of the protective outer cuticle and depends on the periodic synthesis and secretion of proteins, such as collagens and proteolytic enzymes (Johnstone 2000). Below, we discuss how these processes provide clues to the roles played by the ptc, ptr, and hh-r genes in C. elegans. It should be noted that RNAi does not always result in a complete elimination of gene activity. Hence, it is possible that the ptc, ptr, and hh-r genes might have additional functions that will only be uncovered after null mutants corresponding to these genes are obtained.
Molting: The sterol connection
C. elegans is a cholesterol auxotroph that obtains sterols from exogenous sources. Cholesterol starvation or inhibition of lrp-1, which encodes a receptor with similarity to megalin and is essential for cholesterol endocytosis, leads to growth and molting defects (Yochem et al. 1999). Although Drosophila and C. elegans require sterols for viability, their sterol content is so low that it has been suggested that its primary role might be in signaling and not in membrane architecture (Kurzchalia and Ward 2003). In Drosophila, molting is controlled by the steroidal hormone 20-hydroxyecdysone (Riddiford 1993; Thummel 1996). In C. elegans, a sterol-derived hormone regulates dauer larvae formation and molting (Matyash et al. 2004). It was recently shown that the SSD of SCAP can bind directly to cholesterol (Radhakrishnan et al. 2004). Given that members of the SSD protein family collectively promote the transport of proteins, sterols and sterol-modified proteins, such as Hh, this raises the possibility that the PTC and PTR proteins play a similar role in molting (Kuwabara and Labouesse 2002). SCAP is also able to form a homotetramer; if PTC and PTR have the ability to form multimers, this could help to explain the partial functional redundancy of the PTR proteins.
Molting and protein transport
Protein transport and secretion play central roles in molting. Mutations in genes involved in protein secretion and transport, such as rme-8, which is required for receptor-mediated endocytosis, and sec-23, which is involved in ER-to-Golgi transport and encodes a component of COPII (coat protein complex II)-coated vesicles, impair molting (Yochem et al. 1999; Zhang and Kalderon 2001; Roberts et al. 2003). Multiple lines of evidence indicate that the molting defects caused by ptc(RNAi) and ptr(RNAi) could be caused by defects in protein, lipid, or sterol transport. First, the topology of the PTC and PTR proteins is similar to that of transporters, specifically those belonging to the resistance/nodulation/cell division (RND) family of prototypic bacterial multidrug efflux pumps (Paulsen et al. 1996; Kuwabara et al. 2000). Second, ptc-1 and ptr-2 are defective in cytokinesis because they fail to form or maintain a membranous cleavage furrow (Kuwabara et al. 2000; Strickland and Burgess 2004). Third, ptr(RNAi) disrupts alae formation, which is probably dependent on protein exocytosis, as demonstrated for che-14 (Michaux et al. 2000). Finally, disruption by RNAi of a subset of ptr genes perturbs the endocytosis of the yolk protein vitellogenin.
The Hh-r proteins and their relationships with PTC and PTR proteins
The hh-r, ptc, and ptr genes not only promote molting but also affect organismal growth and post-embryonic patterning. Why does depletion of hh-r genes by RNAi produce a phenotype similar to that of ptc and ptr(RNAi)? Here we discuss hypotheses that are open to future investigation. First, given that many of the Hh-r proteins are likely to be secreted, there is a possibility that they could promote cell-to-cell signaling through an interaction with PTC or PTR proteins (Bürglin 1996; Aspöck et al. 1999). If such a receptor/ligand relationship were to exist, which at this time is purely speculative, then genetic arguments postulate that the Hh-r proteins are likely to activate and not inhibit the PTC and PTR proteins because they share similar phenotypes. By contrast, in all other organisms studied so far, Hh binding inhibits Ptc activity and promotes signal transduction via Smo (Ingham and McMahon 2001; Lum and Beachy 2004). Whether the PTC, PTR, and Hh-r proteins could then activate a signaling cascade is also unknown. However, if this were the case, it would be necessary to speculate that PTC and PTR could directly mediate signal transduction or function through a membrane protein other than Smo.
Second, in Drosophila and vertebrates, Hh proteins undergo a maturation process involving autoproteolysis, closely followed by the addition of cholesterol and lipid (Porter et al. 1996b). Similarly, it has been shown that the C. elegans Hh-r protein, WRT-1, undergoes autoproteolysis and has the potential to be modified by lipid and/or cholesterol (Porter et al. 1996a,b; Aspöck et al. 1999). Thus, it is possible that the PTC and the PTR proteins transport the lipid or cholesterol, which is necessary to facilitate these modifications. Third, the PTC and PTR proteins could be involved in the transport or endocytosis of Hh-r proteins. Fourth, the alae defects associated with RNAi of some ptr indicate that they could be mediating exocytosis as does CHE-14. Similar to Disp and Hh, the PTR proteins might be needed to exocytose the Hh-r proteins away from the cell from where they were synthesized (Michaux et al. 2000). In Drosophila, Disp and Hh mutants also share similar mutant phenotypes (Burke et al. 1999).
Finally, it should be stressed that there is no evidence directly linking the functions of the PTC and PTR proteins with those of the Hh-r proteins. It is equally possible that the activities of the PTC and PTR proteins are completely independent of any interaction with the Hh-r proteins. What it is clear, however, is that the Hh-r, PTC, and PTR proteins have functions that are independent of Smo, which might eventually be detected in other organisms. These activities could represent ancestral activities, such as the involvement of PTC-1 and PTR-2 in cytokinesis (Kuwabara et al. 2000; Skop et al. 2004).
Conclusions
The Hh signaling pathway, as deduced from studies in Drosophila and vertebrates, has undergone extensive evolutionary divergence in C. elegans. A number of key members of the pathway are absent, such as Hh and Smo, whereas other members, such as the Ptc, Ptc-related, and Hh-r proteins, are present and have also undergone an expansion in number. Moreover, the C. elegans TRA-1 ortholog of the zinc-finger transcription factor Ci/Gli, which is the terminal regulator of the Hh pathway, is used instead to control nematode sex determination (Hodgkin 1983; Mathies et al. 2004).
It remains unclear whether C. elegans ever had a complete Hh signaling pathway, or whether it was lost at some point in nematode evolution. An acceptable resolution of this question will depend, in part, on the outcome of an open debate regarding the robustness of the ecdysozoa clade assignment, which includes the molting animals Drosophila and C. elegans (Aguinaldo et al. 1997; Wolf et al. 2004). If nematodes, such as C. elegans, are indeed members of ecdysozoa, then there is a strong likelihood that most members of the Hh signaling pathway were present in both Drosophila and nematodes, but were subsequently lost in nematodes.
Methods
BLAST analysis and protein phylogeny
Sequences similarity searches were performed by using BLAST (Altschul et al. 1997). Cluster analysis was performed by using Clustal X, a Windows interface for the Clustal W multiple sequence alignment program (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/) (Thompson et al. 1997). Phylogenetic trees were constructed by using TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) (Page 1996). A total of 1000 bootstraps were run to assess the significance of branching order. C. elegans and C. briggsae sequences are available in Wormbase (http://www.wormbase.org/). Other nematode sequences are available from http://www.tigr.org/tdb/ (The Institute for Genome Research [TIGR]). GenBank accession numbers for non-nematode sequences are as follows: DsHMGCR (P14773), MmHMGCR (XP_127496), HsHMGCR (AAH33692), DmSCAP (NP_788277), MmSCAP (NP_001001144), HsSCAP (Q12770), Mm7DHCR (AAH06854), Hs7DHCR (AAD02816), DmDisp (NP_524734), MmDispA (NP_081142), MmDispB (NP_733481), HsDispA (NP_116279), HsDispB (NP_277045), DmNPC (NP_609357), MmNPC-1 (NP_997125), HsNPC1L1 (AAR97886), HsNPC-1 (NP_000262), DmPTC-1 (AAA28696), MmPTC-1 (NP_032983), MmPTC-2 (NP_032984), HsPTC-1 (NP_000255), HsPTC-2 (NP_003729), DmPTR (NP_610209), MmPTR (XP_109751), and HsPTR (XP_370541).
C. elegans methods
General methods for the handling and maintenance of C. elegans are as previously described (Brenner 1974). The wild-type reference strain is N2 Bristol. The following mutants and transgenic lines were used: rrf-3 (pk1426) (Simmer et al. 2002), him-8 (e1489), and him-5;vit-2::GFP (Grant and Hirsh 1999). The him-8 (e1489); rrf-3 (pk1426) double mutant was generated by using conventional genetic techniques.
RNAi constructs and application
PCR fragments were generated by using primers (sequences available on request from investigators of this study) capable of amplifying ∼0.8 kb of exon-rich DNA sequence, and were cloned into the L4440 RNAi feeding vector (Timmons et al. 2001). Although the ptc and ptr genes encode protein homologs, in most cases it is unlikely that RNAi is eliciting cross-reactivity or secondary transitive effects because the regions affected by RNAi share <65% identity at the nucleotide level. The only possible exceptions include, ptr-3/ptr-12, which share a short (101/120-nucleotide) region of shared identity upstream of the target sequence and the known paralogs, grd-5/grd-10, grd-3/grd-4, and grd-13/grd-14, which share >70% identity at the nucleotide level. All RNAi feeding experiments were performed essentially as described by Timmons et al. (2001).
Acknowledgments
We thank Barth Grant for generously providing the him-5;vit-2::GFP reporter strain, Michel Labouesse and Geraldine Seydoux for sharing unpublished results, and Thomas Bürglin for discussion. We also thank Jonathan Hodgkin, Karen Yook, and members of the Kuwabara laboratory for discussion and comments on the manuscript. SAGE data were produced at the Michael Smith Genome Sciences Centre with funding from Genome Canada. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). P.E.K. is a recipient of a Senior Non-Clinical Fellowship funded by the Medical Research Council.
Footnotes
-
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: B. Grant, M. Labouesse, and G. Seydoux.]
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3935405.
-
↵2 Current address: The Wellcome Trust Sanger Institute, Hinxton, Cambridge, CB10 1SA, UK.
-
↵3 Corresponding author. Email p.kuwabara{at}bristol.ac.uk; fax: 44-117-928-8274.
-
- Accepted June 27, 2005.
- Received March 15, 2005.
- Cold Spring Harbor Laboratory Press