Extrachromosomal Circular DNA of Tandemly Repeated Genomic Sequences in Drosophila
Abstract
One characteristic of genomic plasticity is the presence of extrachromosomal circular DNA (eccDNA). This DNA is found in various eukaryotes from yeast to humans, and its levels are elevated by exposure to carcinogens. eccDNA is heterogeneous in size and composed of chromosomal sequences. In this study we used two-dimensional gel electrophoresis to detect and characterize eccDNA in Drosophila. We found eccDNA throughout the fly's life cycle. These molecules comprise up to 10% of the total repetitive DNA content, and their size ranges from <1 kb to >20 kb. The eccDNA population contains circular multimers of tandemly repeated genes such as histones, rDNA, Stellate, and the Suppressor of Stellate. Multimers of centromeric heterochromatin sequences are included in eccDNA as well. Our findings are consistent with the hypothesis that intramolecular homologous recombination between direct tandem repeats is a favorite mechanism for eccDNA formation. The level of eccDNA increased following MMS treatment of wild-type larvae, consistent with phenomena observed in cultured mammalian cells. This shows mutagen-induced eccDNA formation in the context of the whole organism for the first time. Mutations in the genes okra, mus309, and mei41 did not affect eccDNA under normal conditions or following mutagen treatment, implying that eccDNA formation is different from known pathways of DNA repair.
An intriguing feature of the plasticity of the eukaryotic genome is the formation of extrachromosomal circular DNA (eccDNA), also called small polydispersed circular DNA (spcDNA; Cohen and Lavi 1996; Cohen et al. 1997). eccDNA exists in normal cells but becomes more abundant following exposure of cultured cells to DNA-damaging agents (Sunnerhagen et al. 1989; Cohen and Lavi 1996; Cohen et al. 1997). These extrachromosomal circles, ranging in size from several hundreds of base pairs to several tens of kilobase pairs, are found in cells from various organisms including yeast, Drosophila, Xenopus, mice, hamsters, monkeys, and humans (for review, see Gaubatz 1990). They consist of DNA homologous to a wide variety of chromosomal sequences, and hence are believed to be derived from the chromosomes.
eccDNA is homologous primarily to repetitive chromosomal DNA sequences, in particular those sequences that are organized in the genome as direct tandem repeats (Gaubatz 1990). This, and the fact that many species of eccDNA are organized as multimers of the repeating unit, has prompted us to suggest that they arise from the chromosomes in a mechanism involving homologous recombination between adjacent chromosomal repeats (Cohen and Mechali 2001). Further support for this theory comes from recent experiments in a cell-free system of Xenopus eggs. These experiments showed that eccDNA is formed de novo, independently of replication of chromosomal DNA, from a template that contains direct tandem repeats (Cohen and Mechali 2001). However, the details of the mechanism of eccDNA formation remain to be determined.
eccDNA may be a characteristic of normal development. It exists in Xenopus embryos, where it is most abundant in the stage of premidblastulla transition (MBT), and its level declines sharply in subsequent developmental stages (Cohen et al. 1999). Extracts of activated Xenopus eggs produced eccDNA de novo, indicating that the machinery for forming eccDNA resides in the normal egg and is active in the early embryo (Cohen et al. 1999; Cohen and Mechali 2002). eccDNA was also observed in Drosophila embryos; however, its specificity to the embryonic stage was not examined (Stanfield and Helinski 1976; Pont et al. 1987). Although several specific repetitive sequences were identified in the population of fly embryonic eccDNA (Pont et al. 1987; Degroote et al. 1989), it is still not clear which genomic sequences are prone to appear in extrachromosomal circles and what are the prerequisites for their appearance in eccDNA.
The wealth of knowledge on the development of Drosophila and the genetic tools available for it make it a most suitable model for the study of eccDNA in vivo in the context of a whole organism. However, over the past three decades, this study has been limited by the lack of a convenient technique for the isolation and characterization of eccDNA, and required laborious methods of DNA purification and electron microscopy. These methods were not conducive for comparative studies of the abundance, size, sequence content, and organization of eccDNA between different developmental stages, different genetic compositions, or following various experimental manipulations.
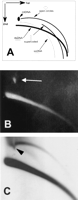
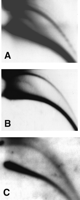
We use a neutral—neutral two-dimensional (2D) gel electrophoresis, which facilitates the study of eccDNA (Cohen and Lavi 1996; Cohen et al. 1997). In this technique, DNA molecules are separated according to both their size and structure so that a population of molecules sharing the same structure, but of heterogeneous molecular mass, generates a continuous arc. Thus, arcs typical of supercoiled molecules, open circles, and linear molecules can be distinguished following hybridization with total genomic DNA or with specific probes (Fig. 1A). The structural identity of the DNA in each arc was determined by electron microscopy (Cohen and Lavi 1996) and verified by molecular techniques (Cohen and Lavi 1996; Cohen et al. 1999; Cohen and Mechali 2001, 2002). This enables the identification of circular molecules within a relatively small sample of total DNA and facilitates structure—function studies of eccDNA.
Two-dimensional gel analysis of extrachromosomal circular DNA in Drosophila embryos. (A) A schematic outline of the 2D gel electrophoretic patterns of genomic DNA generated by populations of linear and circular molecules (Cohen and Lavi 1996). Each arc consists of molecules sharing the same structure, but differing in mass. This analysis allows discrimination between double-stranded DNA (dsDNA), single-stranded DNA (ssDNA), relaxed (open) circular molecules, and supercoiled molecules. It also enables detection of the mitochondrial DNA (mtDNA). (B) Ethidium bromide staining of a 2D gel analysis of genomic DNA from early Drosophila embryos reveals an arc of the chromosomal linear DNA and a spot of the mtDNA (19.5-kb circular DNA, white arrow). (C) Hybridization with total genomic DNA probe reveals an arc of relaxed circles in addition to the massive arc of linear DNA. Note that the large quantity of mtDNA caused a local shift in the migration of the arc of the circular molecules (arrowhead), which extends beyond the mtDNA.
Here we report the beginning of a systematic analysis of eccDNA in Drosophila. We find that the abundance of eccDNA is changed in a defined pattern throughout the life cycle of Drosophila. The population of eccDNA consists of multimers of various tandemly repeated genomic sequences including coding genes and heterochromatic satellite DNA. We further show that treatment with the mutagen MMS increases the level of eccDNA in larvae, in a mechanism that may not be directly linked to known pathways of DNA damage repair.
RESULTS
Drosophila Embryos Contain Abundant eccDNA Consisting of Relaxed Circular Molecules
Total genomic DNA was purified from early embryos (pre-MBT stage) and separated on a 2D gel. Ethidium-bromide staining revealed only the linear DNA and a spot corresponding to the large amounts of mitochondrial DNA deposited into the oocyte (Fig. 1B). Hybridization with total Drosophila genomic DNA revealed two typical arcs: a lower massive arc corresponding to the linear DNA and an upper arc of relaxed circles (Fig. 1C). Relaxed circles, which migrate to the same position whether open or closed, were easily detected in embryos from each of three wild-type strains examined: Canton-S, Oregon-R, and π2. An additional faint arc of single-stranded linear DNA is observed between the linear double-stranded DNA and the eccDNA, and in its low mass range, it crosses the double-stranded DNA (Figs. 1A and 2C). It is sensitive to S1 nuclease (Fig. 2C), and its identity was further validated in previous reports (Cohen and Mechali 2001, 2002).
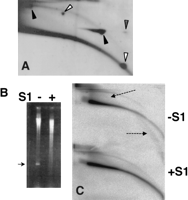
eccDNA is made of relaxed circles. (A) DNA from early embryos, mixed with pUC18-derived plasmids (of 3.3 and 11.2 kb), was separated on a 2D gel. The blot was hybridized with total genomic probe and then with the plasmid probe (pUC18). Superposition of the two autoradiograms shows comigration of the Drosophila eccDNA arc with the relaxed forms of each plasmid (solid arrowheads). The supercoiled plasmid forms migrated to positions below the eccDNA arc (open arrowheads). Stippled arrowhead marks a supercoiled form that was nicked during the first dimension and migrated as a relaxed form in the second dimension (Cohen and Lavi 1996). (B,C) eccDNA is resistant to S1 nuclease. Total genomic DNA from Drosophila early embryos was mixed with M-13-derived single-stranded DNA plasmid and treated with S1 nuclease. A sample was taken before adding the enzyme (-S1), and the other sample after incubation of 5 min with S1 nuclease. (B) The ethidium bromide staining of the first dimension. Arrow indicates the single-stranded marker that disappeared following S1 nuclease digestion. (C) The hybridization pattern of total genomic probe. eccDNA remained unchanged, but single-stranded genomic DNA (dashed arrows) disappeared.
To validate the identity of the DNA that migrated on the nonlinear arc, two circular plasmids were loaded onto the same lane as the embryonic DNA. Hybridization with a plasmid and genomic DNA showed that eccDNA comigrated with the relaxed forms of the plasmids, while the supercoiled forms migrated below the arc of relaxed circles (Fig. 2A). To further confirm the relaxed nature of eccDNA, we treated the embryo DNA with S1nuclease. Supercoiled circles are expected to be nicked by the enzyme and alter their migration position on a 2D gel, while the migration of relaxed circles will remain unchanged. We found the S1nuclease digestion did not affect the migration of eccDNA (Fig. 2C), but a single-stranded marker that was mixed with the embryo DNA disappeared (Fig. 2B) and the linear DNA suffered a certain level of degradation (Fig. 2B,C). This indicates that the eccDNA we detect on our 2D gels is made of relaxed circles.
The strong hybridization signal of eccDNA, which is undetectable by ethidium bromide, can be explained by the fact that because of sequence complexity, not all genomic sequences are equally represented in the hybridization pattern with a total genomic probe. Because of their abundance, repetitive sequences within the probe are more likely to anneal with their homologous repetitive sequences in the genome as compared with unique sequences. Therefore, the pattern of hybridization with total genomic probe represents mainly the repetitive sequences detected. On the other hand, ethidium bromide stains all DNA molecules equally. Thus, our results indicate that repetitive sequences are overrepresented in the eccDNA population compared with a random genomic sequence. This is further demonstrated below with the use of specific sequences as probes.
To obtain an estimate of the abundance of eccDNA, we quantified by Phosphorimager the intensities of the arc of linear DNA and the arc of eccDNA of each sample, and expressed the intensity of the eccDNA arc as a fraction of the sum of the intensities of the two arcs. In spite of its limited accuracy, this analysis revealed that in DNA from pre-MBT embryos of all strains examined, eccDNA comprises 5%—10% of the total hybridization signal, which indicates that 5%— 10% of the total genomic repetitive DNA is circular. Similar values of eccDNA were detected when specific repetitive sequences (as in Figs. 4, 5, 6, 7 below) were used as probes. For example, eccDNA comprised up to 7% and 5% of the 1.688 g/cm3 and the 5S rDNA signals, respectively.
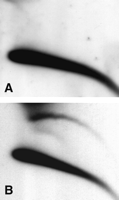
Extrachromosomal circular multimers of 5S ribosomal DNA. Total DNA from adult flies was analyzed on 2D gel, blotted, and hybridized with a cloned copy of the Drosophila 5S ribosomal gene. Circular multimers form a ladder of discrete spots (arrow).
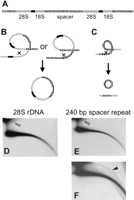
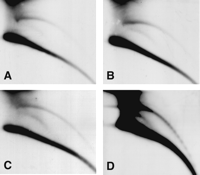
Circular DNA from the ribosomal gene cluster. (A) A schematic outline of the organization of ribosomal genes. Shown are the 28S and 18S genes (gray), the intergenic nontranscribed spacer (stippled), and the internal transcribed spacer (black). The lengths of the genes and the spacers are not drawn to scale. (B) Looping out between spacers or rDNA genes from different clusters yields large circles that include rDNA genes and spacer sequences. (C) Looping out within the short repeats in the spacer yields smaller circles of spacer repeats. (D) Hybridization of a 2D-gel blot of adult DNA with a 28S rDNA probe reveals large circles only (open arrowhead). (E) Short exposure following hybridization with a cloned 240-bp spacer probe reveals large circles (open arrowhead). (F) Longer exposure of the same blot in E reveals also an arc of smaller circles (solid arrowhead).
eccDNA from tandemly repeated protein coding genes. Hybridization of blots containing adults DNA with probes of (A) histone H3; (B) Stellate; (C) Suppressor of Stellate. Ladders of spots represent circular multimers of the repeating units in panels A—C, which are 5 kb, 1.25 kb, and 2.5—2.8 kb long, respectively. (A,C) DNA from wild-type Canton-S. (B) DNA from wild-type Oregon-R.
eccDNA from satellite sequences. DNA was separated on 2D gels and hybridized with probes of Drosophila satellite DNA. (A) 1.688 g/cm3 (359-bp repeat); (B) dodeca satellite; (C) 1.672 g/cm3 satellite.
Most of the cellular repetitive DNA in Drosophila resides in the heterochromatin, which comprises about one-third of the genome content (Adams et al. 2000). Therefore, we estimate that eccDNA comprises ∼1%—2% of the total DNA content. Indeed, when we loaded several tens of micrograms of Drosophila DNA onto the 2D gels, we could detect the arc of eccDNA even by ethidium bromide. In any case, this value is considerably higher than the 0.03% reported before (Stanfield and Helinski 1976).
The size of the eccDNA molecules ranges from <1kb to >20 kb. This was determined by the migration of known plasmids, which were loaded onto the same gel and served as size markers (e.g., Fig. 2A and see Cohen et al. 1999; Cohen and Mechali 2002), and of mitochondrial DNA, which in Drosophila is a 19.5-kb-long circle (Lewis et al. 1995). The large amount of mitochondrial DNA causes a local shift in the arc of eccDNA, seen upon hybridization with a total genomic probe (Fig. 1C). The arc of eccDNA continues beyond that shift, further into the higher-molecular-weight region, indicating that embryonic eccDNA in Drosophila also contains molecules >19.5 kb. Here, too, the size range that we detect is far higher than values in previous reports, which were based on electron microscopy measurements of CsCl-purified supercoiled DNA (Stanfield and Lengyel 1979; Pont et al. 1987; Degroote et al. 1989; see Discussion).
The Levels of eccDNA Change During the Life Cycle of Drosophila
To examine whether eccDNA occurs in other stages of the life cycle beyond pre-MBT embryos, we analyzed in a similar way DNA samples from early and late embryos, from larvae of first and third instar, and from adult flies.
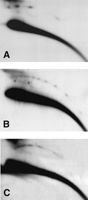
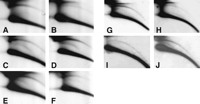
Results for all three wild-type strains examined were essentially similar, and examples are illustrated in Figure 3. The high abundance of eccDNA observed in pre-MBT embryos (Fig. 3A) is followed by a threefold to sixfold decrease of its relative level in late embryos (post-MBT, 6—20 h old) as estimated by quantification of several independent experiments (Fig. 3B). The low level of eccDNA starts to rise again in first-instar larvae (Fig. 3C), returning in third-instar larvae (Fig. 3D) and in adults (Fig. 3E) to the levels observed in the early embryos or sometimes even slightly higher. Hybridization with 5S rDNA (see the detailed description of the hybridization pattern below) reveals a similar pattern of a decrease in the amount of eccDNA between early and late embryos (Fig. 3G,H) and an increase from first-instar to third-instar larvae (Fig. 3I,J).
Profile of eccDNA throughout the life cycle of Drosophila. 2D-gel blots of DNA samples from different stages of the Drosophila life cycle were hybridized with a total genomic DNA probe (A—F) or a 5S rDNA probe (G—J). The panels represent 2D-gel blots of (A,I) pre-MBT embryos, (B,H) late embryos, (C,I) first-instar larvae, (D,J) third-instar larvae, (E) young adult flies (1 day old), and (F) old adult flies (33 days old). The hybridization patterns reveal a decrease in the relative amount of eccDNA during embryogenesis (cf. panels A,B and panels G,H), and a gradual return to the initial levels in the larva and young adult stages. A slight decrease is observed in eccDNA levels in old adult flies compared with young adult flies.
These results indicate that formation of eccDNA is both a maternal (pre-MBT) and zygotic activity. The decrease in the level of eccDNA observed in late embryos was also reported in Xenopus (Cohen et al. 1999), and indicates a developmental regulation on normal eccDNA formation. Note that the faint arc of single-stranded DNA appears in some of our samples. However, we did not detect any correlation between this DNA and a specific developmental stage or defined physiological states.
Extrachromosomal circles of ribosomal DNA accumulate in aged yeast cells (Sinclair and Guarente 1997). To test whether eccDNA accumulates in flies with aging, we compared DNA from adults that were 1day old (Fig. 2E), 7 day old (data not shown), and 33 days old (Fig. 2F). No change in relative eccDNA levels was observed in 1-day-old and 7-day-old adults using 28S rDNA (data not shown) or total genomic DNA as probes. However, in some cases, a decrease of up to threefold was observed in 33-day-old flies (Fig. 2F). We conclude that aging in Drosophila is not accompanied by accumulation of eccDNA; rather, its level may be decreased.
Drosophila eccDNA Contains Multimers of Tandemly Repeated 5S rDNA Genes
In a population of extrachromosomal supercoiled circular DNA isolated from Drosophila embryos by CsCl gradient, Picard and coworkers identified several specific sequences that were all homologous to tandemly repeated chromosomal sequences (Pont et al. 1987; Degroote et al. 1989; Renault et al. 1993). In several cases (5S rDNA genes, satellite 1.688 g/cm3, and the 240-bp spacer repeat of rDNA), the circles were composed of multimers of the repeating unit.
To examine the sequence content and organization of the eccDNA that we detect in Drosophila using the 2D gel approach, which contains primarily relaxed circles, we first used the 5S rDNA as a probe. This gene in Drosophila is 375 bp long and is organized as 165 copies of direct tandem repeats on the right arm of Chromosome 2 (Ashburner 1989). Figure 4 shows the hybridization pattern of total adult DNA with the 5S rDNA probe. In addition to the massive arc of linear DNA, a clear pattern of ladder is observed on the arc corresponding to relaxed circles. This ladder is similar to the pattern of circular multimers of tandem repeats, which were observed in Xenopus (Cohen et al. 1999; Cohen and Mechali 2001). Using circular size markers that were loaded on the same 2D gel, we could determine that each spot on this ladder is an exact multimer of the 375-bp 5S rDNA repeat unit (data not shown), which indicates that this may be the only sequence contained in the eccDNAs. As with the total genomic probe (Fig. 1C), the 5S rDNA probe reveals large circles of >20 kb evident by the continuation of the arc of the circles toward the high-molecular-weight region of the gel. This size is considerably larger than the maximum of 16 repeats (∼6 kb) previously reported for 5S rDNA in extrachromosomal super-coiled circular DNA from Drosophila (Pont et al. 1987). This multimer organization and size range was detected in all strains tested and did not change throughout the life cycle of the fly (data not shown).
To further verify that the ladder is composed exclusively of multimers of the 5S rDNA repeats, we examined whether unrelated sequences are interspersed in them. To this end we digested the DNA, prior to its 2D gel analysis, with HinPI, which cleaves once in the 5S rDNA gene, or with enzymes that do not cleave in the gene (e.g., EcoRI, HindIII, SacI, XbaI). If unrelated sequences were included in the circles that are homologous to 5S rDNA, these circles (in particular, the large ones) would be sensitive to restriction enzymes that do not cut in the 5S rDNA. Although digestion with HinPI abrogated the ladder, the other enzymes did not affect it (data not shown). Thus, the circular molecules that are homologous to the 5S rDNA consist of the 5S gene exclusively without other intervening genomic sequences.
Distinct Classes of Circles Derive From the rDNA Cluster
The 28S and 18S rDNA genes of Drosophila are arranged in tandemly repeating units organized in clusters of 250 and 200 copies on Chromosomes X and Y, respectively (Tartof 1973; Ashburner 1989). Each unit consists of an 8-kb coding region (that includes the 28S and 18S genes) flanked by a nontranscribed spacer of a variable size (4—8 kb in >80% of the cases; Fig. 5A). The spacer itself consists of short repeating elements that share some homology with each other (Wellauer and Dawid 1977; Ashburner 1989).
Previous studies on supercoiled circles in Drosophila embryos reported sequences homologous only to the 240-bp spacer repeat (but not to the other spacer repeats or the 28S and 18S rDNA genes) in that population of molecules (Pont et al. 1987).
If circles were formed by intramolecular homologous recombination, we would predict that sequences homologous to the transcribed 28S and 18S rDNA genes will appear only in large circles of >10 kb, as a result of looping out of one or more whole clusters (at least 8 kb of the transcribed genes plus at least part of the spacer; Fig. 5B). The repeating units within the nontranscribed spacer elements would appear either in smaller circles, resulting from looping out within a single spacer (Fig. 5C), or be included in larger molecules that consist of variable numbers of the transcribed gene cluster together with their adjacent spacers (Fig. 5B).
Hybridization with the 28S rDNA probe revealed a short arc of large circles (Fig. 5D) as expected from looping out of one or more whole gene clusters (Fig. 5B). This hybridization pattern did not change at longer exposure (data not shown). Discrete spots of multimers were not observed on this arc because of the variable length of the spacer, which would cause even circular monomers to be heterogeneous in size. Hybridization with the 240-bp spacer probe revealed large circles, similar in size to those obtained with the 28S rDNA probe after a short exposure (Fig. 5E). However, after a longer exposure we also detected smaller circles ranging in size from <1kb to >10 kb (Fig. 5F) as predicted in the scheme in Figure 5C. The resolution of our 2D gel made it difficult to detect a ladder of multimers of the 240 bp, but in some experiments we did observe it (data not shown).
The large circles that hybridized to the 240-bp repeat probe were sensitive to digestion with enzymes that cleave once in the transcribed region (e.g., HindIII), as expected if in addition to the spacer sequences, these molecules consisted of the transcribed 28S and 18S rDNA genes. The smaller circles were resistant to HindIII cleavage, as expected from multimers consisting of the 240-bp repeat only (data not shown). These findings are in agreement with the hypothesis of formation of extrachromosomal circles homologous to rDNA by recombination between adjacent clusters or within the spacer.
Protein-Coding Genes Are Also Prone to Formation of Open Extrachromosomal Circles
Our results indicate that other genes that are organized in the chromosome as direct tandem repeats, including protein-coding genes, might also be present in the eccDNA, as multiples of their repeating unit. To test this, we used as probes sequences of the following three representative genes: histone H3 (as a marker of the five histone genes), Stellate, and Suppressor of Stellate.
The histone genes of Drosophila are arranged on Chromosome 2 in ∼100—110 direct tandem copies of a 5-kb unit that includes all the five histone genes (H1, H2A, H2B, H3, and H4; Lifton et al. 1978). Picard and colleagues have detected circular monomers of the 5-kb unit of the histone genes in the population of Drosophila circular DNA (Pont et al. 1987). Hybridization of a 2D gel with histone H3, as a representative probe of this repeating unit, revealed four discrete spots corresponding to circles of 5, 10, 15, and 20 kb, as expected from multiples of the repeating unit (Fig. 6A). We presume they also exist in multimers larger >20 kb, but the resolution of our 2D gel cannot detect them. The 20-kb spot is less intense than the 15-kb spot, probably because of inefficient extraction of large circles. Alternatively, if intramolecular homologous recombination was their mechanism of formation, the large circles may be formed inefficiently in vivo because such recombination decreases with the distance (Ringrose et al. 1999).
The X-linked Stellate (Ste) gene encodes a protein with a significant identity to the β subunit of casein kinase 2. The gene is organized in direct tandem repeats in distinct clusters located in the euchromatin as well as the heterochromatin. The euchromatic repeating unit of Ste is 1.25 kb long, and the heterochromatic is 1.15 kb. Both types of repeats are expressed and differ in their 3′ noncoding region (Livak 1990; Shevelyov 1992). The ratio between the number of euchromatic and the heterochromatic types varies in different strains, and the number of the repeats ranges between 20 and 200 (Livak 1984). In the wild-type strain Oregon-R, the vast majority of Ste copies are of the euchromatic type, and their number was estimated to be 200 copies per haploid genome (Livak 1984). Figure 6B shows the hybridization pattern of a blot containing genomic DNA from Oregon-R flies probed with Ste. A clear ladder is observed, and the spots correspond to multiples of 1.25 kb.
The gene Supressor of Stellate [Su(Ste)] is also organized in tandem repeats. Like Ste, the Su(Ste) gene contains an open reading frame homologous β subunit of casein kinase 2. It has two size variants (2.5 and 2.8 kb), both of which are expressed despite being embedded in the constitutive heterochromatin of the Y chromosome. Their number varies in different strains and was estimated to be 80 copies in Oregon-R (Livak 1984). Using a Su(Ste) probe, we detected a ladder of wide rods, rather than round spots, on the arc of the circular molecules that correspond to multiples of 2.5 to 2.8 kb (Fig. 6C). This result was reproducible in several fly strains and indicates that the rods are not exact multiples of either 2.5 or 2.8 kb, but may consist of a mixture of these two variants, in agreement with their heterogeneous organization in the clusters (Kalmykova et al. 1998).
Tandem Heterochromatic Satellite Repeats Can Also Form eccDNA
Whereas the repeated sequences described above are all expressed, a great majority of the tandem repeats in the genome reside in the heterochromatin and consist of noncoding short repeat units. For example (see Table 1), the 1.672-g/cm3 satellite (AATAT tandem repeat) is present in a 3.5-Mb-long stretch of DNA on Chromosome 4 of Drosophila (Dernburg et al. 1996), and the dodeca satellite (an 11- or 12-bp tandem repeat) comprises 1 Mb at the pericentromeric region of Chromosome 3 (Abad et al. 1992). A somewhat longer heterochromatic tandem repeat (359 bp) is represented by the 1.688-g/cm3 satellite, which constitutes 11 Mb at the centromeric region of the X chromosome (Dernburg et al. 1996). We found sequences homologous to each of these three heterochromatic repeats in the arc of eccDNA following 2D gel analysis of DNA from the three wild-type strains examined and at various developmental stages (Fig. 7; data not shown). The hybridization pattern of the 1.688-g/cm3 satellite revealed a ladder of spots corresponding by size to multimers of the 359-bp repeating unit comparable to that of the 5S rDNA (Fig. 7A). We presume that similar ladders of multimers exist for the shorter repeats of the dodeca satellite (Fig. 7B) and the 1.672-g/cm3 satellite (Fig. 7C), but because our 2D gel cannot resolve them, they appear as continuous arcs.
Chromosomal Tandem Repeats in eccDNA of Drosophila
Taken together, our results indicate that heterochromatic coding and noncoding tandem repeats are prone to formation of extrachromosomal relaxed DNA circles as are euchromatic repeats. In all cases the predominant organization of the circles is as multiples of the repeating unit.
Dispersed Repeats Are Not Represented in Drosophila eccDNA
An emerging conclusion from this and previous studies is that arrangement in tandem direct repeats is a prerequisite for formation of eccDNA. To further test this assertion, we examined whether repeated sequences that are dispersed in the chromosomes are also found in eccDNA. The P transposable element served as an example. In the Drosophila π2 strain, the transposition of this element is repressed and it is present in the genome in some 20—30 copies dispersed on all chromosomes (FlyBase 1999). Hybridization with a P-element probe that recognizes essentially all copies of P in this genome (including the defective ones; see Methods) revealed no sequences homologous to it in the arc of eccDNA from this strain even following long exposure (Fig. 8A). Rehybridization of the same blot with total genomic DNA probe showed that this DNA preparation does contain eccDNA (Fig. 8B). Similarly, the transposable I element was absent from the circular DNA population from Drosophila embryos (Pont et al. 1987). In Xenopus embryos, too, a dispersed repeat was not present in the eccDNA population (Cohen et al. 1999). This finding further supports the idea that tandem repeats are prone to formation of eccDNA (Cohen and Mechali 2001), whereas dispersed repeats form such circles to a much less extent, if any.
Absence of eccDNA homologous to the dispersed P element. (A) A 2D-gel blot of wild-type (π2) adult DNA, hybridized with a cloned 5′ region of the P transposable element, reveals only the linear arc. (B) Rehybridization of this blot with a total genomic DNA probe reveals both the arc of eccDNA and the arc of linear DNA, indicating that the absence of the eccDNA arc in panel A reflects the absence of P-element sequences within the eccDNA population.
Mutants in Several Genes Implicated in Genomic Integrity Display a Normal Pattern of eccDNA
The results described heretofore and the mechanistic evidence from the Xenopus cell-free system (Cohen and Mechali 2001) indicate that intramolecular homologous recombination between repeats may account for the formation of eccDNA. In Saccharomyces cerevisiae, such a mechanism for the generation of rDNA circles was demonstrated genetically. Among the yeast genes that are involved in this mechanism are SGS1 (a helicase of the recQ family) and the RAD52 pathway (Park et al. 1999). We asked whether Drosophila genes that are involved in mitotic recombination play a role in eccDNA formation, and chose to examine eccDNA in mutants of the Dmblm and okra genes. Dmblm (mus309) is the Drosophila homolog of the human blm gene. It encodes a helicase of the RecQ family (as does SGS1), and it is implicated in DNA double-strand break repair (Kusano et al. 2001). okra is the Drosophila homolog of RAD54, and is involved in meiotic and X-ray-induced mitotic recombination (Kooistra et al. 1997). We examined eccDNA in mutant adult in comparison with heterozygous flies (rather than with a laboratory wild-type strain) to overcome the variability in the amount of eccDNA between different strains or in different genetic backgrounds.
The heteroalleles of the two null mutations okrAG/okrA17-11 and the corresponding hemizygous combinations okrAG/Df(2L)JS17 and okrA17-11/Df(2L)JS17 contained levels of eccDNA similar to those of the heterozygotes okrA17-11/CyO and Df(2L)JS17/CyO as determined by hybridization with total genomic probe (Fig. 9A-C). They also formed normal exact multimers of 5S rDNA (Fig. 9D) and histones (data not shown). Likewise, we did not detect any effect of heteroallelic and hemizygous combinations of mus309 mutations on the amount or multimer organization of eccDNA.
Mutations in okra do not affect eccDNA. DNA from adult flies carrying the following genotypes was separated on 2D gel: (A,D) okrA17-11/okrAG; (B) okrA17-11/Df(2L)JS17; (C) okrA17-11/CyO. The blots in panels A—C were hybridized with total genomic DNA probe; in panel D, a 5S rDNA probe was used. No change in the relative amount of eccDNA was observed in the mutant genotypes compared with the heterozygous one. The circular 5S rDNA multimer organization in the mutants is similar to that observed in the wild type (cf. panel D to Fig. 4).
The X-linked gene mei41 is the Drosophila homolog of ATR and is implicated in various processes of maintenance of genome integrity. Its null mutation mei41D3 is homozygous viable and exhibits high sensitivity to a wide range of physical and chemical mutagens (Boyd et al. 1976). However, homozygous females or hemizygous males did not exhibit abnormal amounts or patterns of eccDNA (data not shown).
Mutants in mei9 (RAD1 homolog, implicated in end joining and other types of repair; Sekelsky et al. 1995), mus304 (a checkpoint gene implicated in genomic integrity; Brodsky et al. 2000), and grapes (checkpoint 1, interacts with mei41; FlyBase 1999; Sibon et al. 1999) did not affect eccDNA population. These results indicate that none of the genes tested is involved in the formation or maintenance of eccDNA in adults, and as described below, not in the larval stages either.
The Mutagen Methyl Methansulphonate Increases the Levels of eccDNA in Vivo
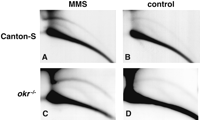
The levels of eccDNA are elevated following exposure of cultured mammalian cells to various agents that cause DNA damage (Sunnerhagen et al. 1989; Cohen and Lavi 1996; Cohen et al. 1997). We asked whether this occurs also in vivo when the whole organism is exposed to a drug that causes DNA damage. Methyl methansulphonate (MMS) is an alkylating agent that causes double-strand breaks. First-instar wild-type (Canton-S) larvae were fed with MMS, and 24 h later their DNA was purified. We detected a twofold to threefold increase in the level of eccDNA in the treated larvae compared with the untreated control when hybridizing with a total genomic DNA probe (Fig. 10A,B) or with the specific probes mentioned above (data not shown). Such increase was found in more than 10 independent experiments in which larvae of Canton-S, π2, and additional laboratory strains were treated. This result demonstrates for the first time that eccDNA is increased in response to DNA damage in the context of the whole organism.
Effect of MMS treatment on eccDNA. (A,C) Drosophila first-instar larvae were fed with 0.1% MMS, and 24 h later DNA was purified and separated on 2D gels that were blotted and hybridized with total genomic Drosophila DNA. (B,D) DNA from nontreated control larvae. (A,B) DNA from wild-type (Canton-S) larvae. (C,D) DNA from mutant okrAG/okrA17-11 larvae (okr-/-).
The Drosophila mutants mentioned above—okra, Dmblm, and mei41—are all defective in genes that might play a role in recombination and DNA repair pathways, and exhibit sensitivity to MMS, manifested as inability to reach adulthood following feeding of larvae with this drug (Boyd et al. 1976; Kooistra et al. 1997; Sibon et al. 1999; Kusano et al. 2001). However, none of the mutants affected the normal increase in eccDNA levels in response to MMS feeding (Fig. 10C,D; data not shown). The multimer organization of 5S rDNA circles was not affected either following MMS treatment of all three mutants (data not shown).
We conclude that although the okra, Dmblm, and mei41 genes play an important role in response to DNA damage, none of them participates in the control of eccDNA formation under normal conditions or following MMS-induced DNA damage.
DISCUSSION
We have begun here a systematic investigation of a phenomenon that reflects plasticity of the Drosophila genome, namely, the formation of extrachromosomal relaxed DNA circles. We show, for the first time, a pattern of change in the levels of eccDNA throughout the normal life cycle, from embryo to adult.
Normally, circular DNA is expected to be present in the cell in a supercoiled form. However, supercoiled DNA was not detected in any of our Drosophila DNA preparations except for one case in DNA from embryos at the MBT stage from mei41D3 homozygous females (data not shown). The significance of this finding is not clear yet, but it ensures that our DNA extraction procedure can successfully purify supercoiled circles and probably does not introduce nicks into the circular DNA. Similarly, although supercoiled circles were not detected in eccDNA from Xenopus embryos, the same extraction method recovered supercoiled plasmids, which were preinjected into fertilized eggs, along with genomic DNA (Cohen et al. 1999). Thus, the apparent absence of supercoiled molecules from our preparations of Drosophila DNA is likely not an artifact but indicates that the vast majority of eccDNA in Drosophila consists of relaxed circular molecules. Possibly, supercoiled molecules, which were previously reported in Drosophila (Stanfield and Helinski 1976; Pont et al. 1987; Degroote et al. 1989), correspond to a minor fraction of the eccDNA that is below the detection limits of our assay. Indeed, isolation by successive CsCl gradients of DNA from a large amount of embryos (∼80 g; Pont et al. 1987), was required to detect super-coiled circles, whereas we detect eccDNA in preparations of 100 mg of embryos or less. However, the relaxed circles are lost during DNA isolation by CsCl gradient, and as they comprise the majority of eccDNA population, this isolation method greatly underestimates the amount of circles in the genome. It may be also nonrepresentative with respect to the size of the DNA circles, because the supercoiled molecules previously reported consisted primarily of small circles (i.e., <2.5 kb and at most 5 kb long; Stanfield and Helinski 1976; Degroote et al. 1989), whereas in the present work we detected much larger eccDNA (even circles >19.5 kb).
The scarcity of supercoiled DNA molecules both in Drosophila and Xenopus could be explained by the results of kinetics studies on eccDNA formation performed with Xenopus egg extracts. That work shows that eccDNA is first formed as relaxed circles and the supercoiled forms appear only later (Cohen and Mechali 2001). However, in vivo, rapid turnover of the relaxed circles into noncircular forms may prevent their availability for conversion into a supercoiled form (Cohen et al. 1999). Alternatively, supercoiled molecules might be formed normally but immediately subjected to rapid metabolism, and thus would not accumulate in the cell. In any case, the reports on Xenopus and our present results detect the steady-state amount of eccDNA in the cells, and indicate that the half-life of the circles is probably very short.
We found that upon hybridization with a total genomic probe (in which repetitive sequences are more prone to anneal to their homologs), eccDNA shows a strong signal that constitutes up to 10% of the total hybridization signal. This indicates that the relative amount of repetitive DNA is higher in eccDNA than in the genome. This conclusion is supported by the easy detection of eccDNA homologous to various tandemly repeated chromosomal sequences. As summarized in Table 1, these sequences are variable: they can be either heterochromatic or euchromatic; they may be expressed (e.g., rDNA, histones, Ste, and Su(Ste)) or noncoding, such as the satellite elements; and they may be very short (repeats of several base pairs) or a few kilobases long (e.g., histones and rDNA genes). Dispersed repeats in the genomes of Drosophila (P element; Fig. 8) and Xenopus (JCC31; Cohen et al. 1999) were not detected in eccDNA. We predict that any tandem repeat in the genome can serve as a template for the cellular-circle-forming machinery regardless of its length or sequence content.
Without exception, whenever the size of the repeated sequence examined was long enough to be resolved by our 2D gels (>350 bp), we found that the eccDNA population comprised circles that were exact multimers of the repeat (Figs. 4, 6, and 7), yielding ladders of circles of a wide size range, from <2 kb to >20 kb. These results are consistent with the multimer organization of eccDNA, which was previously demonstrated in Drosophila (5S rDNA, satellite 359 bp, and 240-bp ribosomal spacer repeat; Pont et al. 1987; Degroote et al. 1989), in humans (α-satellite; Kiyama et al. 1986, 1987), and in Xenopus (satellite 1; Cohen et al. 1999). Experiments with Xenopus egg extracts showed that eccDNA was generated de novo from a cosmid template that contained tandem copies of a 2-kb fragment. The circles consisted of exact multiples of the repeating 2-kb element, whereas the unique sequences of the cosmid were not included. This indicated that the tandem organization was a prerequisite for circle formation (Cohen and Mechali 2001). The process occurred independently of replication of chromosomal DNA, lending further support to the idea that intra-molecular recombination (“looping out,” as illustrated in Fig. 5B,C) is involved in formation of eccDNA. Looping out has been implicated in the phenomenon of class switch rearrangements of immunoglobulin genes (von Schwedler et al. 1990) and in the formation of extrachromosomal rDNA circles in yeast (Park et al. 1999).
Because eccDNA is a heterogeneous population of circular molecules, we cannot rule out the possibility that it contains subgroups of molecules that were formed by other mechanisms like retrotransposition. Alternative mechanisms for the formation of circles from chromosomal DNA could include nonhomologous end joining and single-strand annealing. However, as discussed in detail previously (Cohen et al. 1999; Cohen and Mechali 2001), both require chromosomal breakage as a first step and cannot explain the normal appearance of relatively large amounts of eccDNA.
Note that there is no evidence for a link between eccDNA and apoptosis. On the contrary, eccDNA appears in Xenopus during early embryogenesis, and apoptosis does not occur before gastrulation (Hensey and Gautier 1997). In addition, FACS analysis of cultured mouse cells that exhibited high levels of eccDNA showed that they were not apoptotic (S. Cohen, unpubl.).
In addition to eccDNA being an integral feature of the normal plasticity of the animal genome, its level is increased in response to exposures of cultured mammalian cells to various agents that cause DNA damage and cell cycle arrest including MNNG, DMBA, hydroxyurea, and cycloheximide (Sunnerhagen et al. 1989; Cohen and Lavi 1996; Cohen et al. 1997). Cultured Drosophila cells also exhibited elevated levels of eccDNA after long exposure (14 h) to puromycin and cycloheximide (Stanfield and Helinski 1976). Although these studies were conducted on cultured cells, they have indicated that eccDNA might serve as a short-term marker of carcinogen exposure in vivo. Here we show for the first time that DNA damage caused by MMS increased eccDNA levels in vivo, in the context of a whole organism. It will be interesting to determine whether this increase is similar in all body tissues or whether some tissues exhibit a different response to the treatment compared with others (e.g., the imaginal discs, which contain dividing cells, in comparison to nondividing larval tissues).
It remains to be seen whether the rise of eccDNA level in response to MMS treatment is specific to this mutagen, or is a general response to various types of DNA damage and possibly to other stresses. As mentioned above, in cultured mammalian cells other drugs also cause an increase in eccDNA level. We therefore speculate that similar increases will occur after exposure of Drosophila to different stresses. Thus, our findings strongly support the possible use of eccDNA as a marker for exposure of a whole organism to carcinogens.
To begin a study of the genetic control on eccDNA formation and enhancement by MMS, we tested eccDNA in six mutants, which are defective in mitotic recombination and DNA repair. However, none of them exhibited any appreciable change in the amount of eccDNA or its multimer organization, under normal conditions or following MMS treatment (Figs. 9 and 10). Thus, eccDNA formation and its rise in response to MMS treatment may involve other mechanisms than those affected by these mutants. A similar case of a mutation that confers high recombination rates and genomic instability without affecting eccDNA was recently reported in S. cerevisiae. The hyperrecombination mutant hpr1Δ has a reduced life span and increased genomic instability. Although it enhances the rate of recombination within the rDNA repeats, the level of its extrachromosomal rDNA circles remains unchanged (Merker and Klein 2002).
Although eccDNA is abundant in many eukaryotes, its physiological significance remains obscure. It may be difficult to ascribe a common role to eccDNA species as diverse as coding genes and short satellites. Whether they have different roles or whether some play no role and are generated as by-products of a mechanism that evolved for other purposes still remains to be investigated. It is conceivable that eccDNA might explain the phenomenon of “orphons”—genetic elements, derived from tandem repetitive genes, which are located outside of the cluster (Childs et al. 1981), or play a role in the metabolism and plasticity of tandem repeats.
METHODS
Preparation of Genomic DNA
Total genomic DNA was prepared by rapid homogenization of 50—100 mg of Drosophila embryos/larvae/adults in 30 mM EDTA, 1% SDS, 0.5% Triton X-100, and 0.3 M NaCl, with subsequent incubation at 50°C for ∼4 h with 1mg/mL proteinase K. The DNA was extracted with equal volumes of phenol and phenol:chloroform and precipitated with ethanol or isopropanol. The DNA was resuspended in 1× SSC and digested with 0.2 mg/mL RNase A at 37°C for 1h, followed by phenol:chloroform extraction and ethanol precipitation. The precipitated DNA was resuspended in TE and was ready for further manipulations. Usually the yield was sufficient for 5—10 2D gels.
Neutral—Neutral 2D Electrophoresis
Separation of DNA in neutral—neutral 2D gels was performed according to Brewer and Fangman (1987), with modifications as described (Cohen and Lavi 1996; Cohen et al. 1997). Briefly, the DNA was first separated on 0.4% agarose at 1V/cm in 1× TBE, and the gel was rinsed in 1× TBE containing 0.3 μg/mL EtBr. The lane of choice was cut and placed on a clean gel support at 90° to the direction of the first electrophoresis. The lane was cast with 1% agarose containing 0.3 μg/mL EtBr, and was electrophoresed in 1× TBE in the presence of 0.3 μg/mL EtBr. The first dimension was run overnight at 1V/cm, and the second dimension for 5 h at 4 V/cm, both at room temperature.
Blotting and Hybridization
The gels were agitated in 0.25 M HCl for 30 min, rinsed in water, and agitated again in 0.4 M NaOH for at least 30 min. Southern blotting was performed with Hybond N+ nylon membranes (Amersham). Oligonucleotide probes were end-labeled with polynucleotide kinase (Promega). Other probes were labeled by a random priming kit (Biological Industries). Radiolabeled DNA was detected by autoradiography, and the signal was quantified with a Phosphorimager (Fugi FLA 2000) by using the Tina 2.10g software. The signal obtained from the arc of linear plus the arc of eccDNA is referred to as the signal of the total DNA. We calculate the percentage of the signal obtained from the eccDNA arc relative to the signal of the total DNA.
As probes for 5S rDNA, 28S rDNA, 240-bp rDNA spacer, histone H3, and 1.688 satellite (359-bp satellite), we used PCR fragments that were prepared according to the published corresponding sequences (accession nos. X01082, M21017, X02211, X14215, and AE002811, respectively). The PCR products were sequenced and verified for accuracy. The oligonucleotide (ACTGGTCCCGT)3 served as a probe for dodeca satellite, and the oligonucleotide (AATAT)5 for the 1.672 satellite. The Stellate probe was a 700-bp insert of p14Sa4 containing the Stellate-specific Sau3A fragment. The Suppressor of Stellate probe was a 350-bp insert of pBKS-Ysp containing the Y-chromosome-specific fragment of Su(Ste). The P-element probe was a 500-bp fragment from the 5′ end of a cloned P element (Carnegie Plasmid; Rubin and Spradling 1983). Its sequences are present in intact and defective P elements (Ashburner 1989).
S1 Nuclease Digestion
Embryo DNA (∼5 μg) was mixed with 800 ng of M13-derived single-stranded plasmid (pcs11) and S1 nuclease reaction buffer. An aliquot was removed as a control before adding 20 units of S1nuclease at room temperature for 5 min of incubation. The reaction was terminated with 10 mM EDTA. DNA was phenol:chloroform-extracted and ethanol-precipitated before 2D gel analysis.
Fly Techniques
All Drosophila melanogaster strains were maintained, and crosses were performed, at 25°C in shell vials supplemented with cornmeal—molasses Drosophila medium. The wild-type strains used were Canton-S, Oregon-R, and π2 (a wild-type P-cytotype strain that contains several dozens copies of the transposable P element; FlyBase 1999). All mutations and balancer chromosomes are described in FlyBase (FlyBase 1999). The mutant strains used were mus309D2/TM3; mus309D3/TM6; Df(3R)T-7/TM3 (containing a deletion of mus309/Dmblm); okrAG/CyO; okrA17-11/CyO; Df(2L)JS-17/CyO (containing a deletion of okra); mei41D3/C(1) DX; grp06034/CyO; mei9A2/FM7; and mus304D1/TM3; mus304D3; Df(3L)W4/TM6 (containing a deletion of mus304).
DNA was extracted from homozygous mutants whenever these were viable as adults. The chromosomes carrying mus309D2, mus309D3, okrA17-11, and mus304D1 all harbored additional unrelated lethal mutations. Therefore, in these cases we used heteroallelic combinations or hemizygotes over the corresponding chromosomal deletion. In addition, heterozygotes (over the corresponding balancer) were used.
To identify the genotype of the larvae, all balancers were replaced with corresponding balancers carrying a GFP transgene that is expressed in the larval stage (FlyBase 1999). This enabled distinction between normal (GFP-positive) and mutant (GFP-negative) larvae.
All mutations used in this study confer sensitivity to MMS. This is manifested as lethality and inability to reach adulthood after feeding larvae with 0.05%—0.1% MMS, as reported previously (FlyBase 1999) and verified by us.
Staging Drosophila
For collection of embryos, flies were allowed to lay eggs at 25°C for 1h on agar plates supplemented with live yeast paste. Then parents were removed and embryos were allowed to develop for an additional 1.5 h (for pre-MBT stage), or ∼12—16 h (for late embryos) before harvesting. For collection of larvae, eggs were laid overnight as above, then parents were removed and embryos and larvae were allowed to develop at 25°C for an additional 24 h (first instar) or 72—96 h (third instar) before harvesting.
MMS Treatment of Larvae
Flies were allowed to lay eggs overnight on agar plates, and were subsequently removed. Plates containing the developing embryos were left at 25°C for an additional 24 h (to obtain first-instar larvae). Then 1.2 mL of 0.1% methyl methanesulphonate (MMS, Aldrich Chem. Co.) dissolved in water, or of water only (control), was supplemented to the plate, and absorbed by the agar and yeast paste. Larvae were fed on this medium for 24 h; then they were harvested for DNA extraction.
Acknowledgments
The fly strain okrAG/CyO was obtained from T. Schupbach. Strains okrA17-11/Cyo, Df(2L)JS-17/CyO, and mus309D2/TM3 were obtained from J. Eeken. Strains mus309D3/TM6 and mei9A2/FM7 were provided by J. Sekelsky. Strain Df(3R)T-7/TM3 was provided by J. Szabad. All other fly strains were obtained from the Drosophila stock centers in Bloomington, Indiana and Umea, Sweden. p14Sa4 was a gift from A. Tulin, and pBKS-Ysp was a gift from F. Bantignies. We thank anonymous referees for constructive comments on this manuscript. S.C. was supported by fellowships from the Israel Cancer Research Fund, the Israeli Ministry of Absorption, and the George S. Wise foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: T. Schupbach, J. Eeken, J. Sekelsky, J. Szabad, A. Tulin, and F. Bantignies.]
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.907603.
-
↵2 Corresponding author. E-MAIL scohen{at}post.tau.ac.il; FAX 972-3-6409407.
-
↵1 Present address: Department of Biological Regulation, Weizmann Institute of Sciences, Rehovot 76100, Israel.
-
- Accepted March 14, 2003.
- Received October 13, 2002.
- Cold Spring Harbor Laboratory Press