Detection of Peptides, Proteins, and Drugs That Selectively Interact With Protein Targets
- Ilya G. Serebriiskii1,
- Olga Mitina1,2,
- Elena N. Pugacheva1,
- Elizaveta Benevolenskaya3,
- Elena Kotova1,
- Garabet G. Toby1,4,
- Vladimir Khazak5,
- William G. Kaelin3,
- Jonathan Chernoff1, and
- Erica A. Golemis1,6
- 1Division of Basic Science, Fox Chase Cancer Center, Philadelphia, Pennsylvania 19111, USA; 2Department of Molecular Biology and Medical Biotechnology, Russian State Medical University, Moscow, Russia; 3Dana-Farber Cancer Institute, Boston, Massachusetts 02115, USA; 4Cell and Molecular Biology Group, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania 19104, USA; 5Morphochem, Inc., Monmouth Junction, New Jersey 08852, USA
Abstract
Genome sequencing has been completed for multiple organisms, and pilot proteomic analyses reported for yeast and higher eukaryotes. This work has emphasized the facts that proteins are frequently engaged in multiple interactions, and that governance of protein interaction specificity is a primary means of regulating biological systems. In particular, the ability to deconvolute complex protein interaction networks to identify which interactions govern specific signaling pathways requires the generation of biological tools that allow the distinction of critical from noncritical interactions. We report the application of an enhanced Dual Bait two-hybrid system to allow detection and manipulation of highly specific protein–protein interactions. We summarize the use of this system to detect proteins and peptides that target well-defined specific motifs in larger protein structures, to facilitate rapid identification of specific interactors from a pool of putative interacting proteins obtained in a library screen, and to score specific drug-mediated disruption of protein–protein interaction.
[Supplemental material is available online at http://www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: A. Taliana, M. Russell, M. Berman, and R. Finley.]
Since its inception (Fields and Song 1989), the two-hybrid system has been utilized in increasingly complex strategies to analyze interactions between proteins of biological interest and known or novel cognate partners including other proteins, RNA sequences, pharmacological agents, and peptides (for review, see Serebriiskii et al. 2001). More recently, a number of groups have exploited the potential of two-hybrid systems as a tool for understanding protein interactions on a genome-level scale, with pilot studies involving elucidation of large sets of protein interactions developed in Saccharomyces cerevisiae (Schwikowski et al. 2000; Ito et al. 2001) providing a model for ongoing work in higher eukaryotes. Given the increasing realization that protein interaction networks involve the interaction of discrete signaling molecules with multiple partner proteins in different biological circumstances, accurate description of the function of a given protein now implicitly involves dissection of its interaction domains, ranking of its interaction affinity with each of its partners, and determination of physiological conditions under which each pair of proteins preferentially interacts. These determinations pose significant technological hurdles in high-throughput efforts.
We have described previously a proof-of-concept experiment for a two-hybrid Dual Bait system that provides internal controls for interaction specificity, and could theoretically be used to selectively compare the interaction of a protein with more than one partner molecule (Serebriiskii et al. 1999). Building from this preliminary study, we have now developed a complete system of reagents that can be used to score interaction of one transcriptional activation domain (AD)-fused prey protein with either of two DNA-binding domain (DBD)-fused bait proteins over a range of different interaction affinities. In three different library screening applications, we demonstrate that these reagents can be used to identify proteins or peptides that target short sequence elements of biological importance within a larger protein structure. We further demonstrate that the system can be used in a bait swap application for rapid secondary screening to sort multiple library hits into subgroups most likely to be reproducible and physiologically relevant. Finally, we describe the use of the reagents in a subtractive two-color visualization procedure that can discriminate specific from nonspecific drug-induced inhibition of protein interactions. These studies, together with our other work involving use of the system to build enhanced specificity derivatives of signaling proteins engaged in complex interactions, indicate the Dual Bait is a useful tool in dissection of complex cellular regulatory machinery.
RESULTS AND DISCUSSION
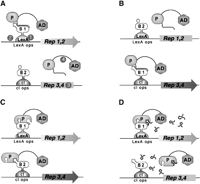
In the Dual Bait two-hybrid system (Fig.1), the use of two parallel bait-reporter systems allows simultaneous and comparative assessment of the interaction of a protein with two discrete partners. The bacterial LexA protein and the bacteriophage λ cI protein provide DBDs for the two baits. Each bait directs the transcription of two matched reporter genes, one colorimetric and one auxotrophic, such that lacZand LEU2 are transcriptionally responsive to LexA fusions, andGusA and LYS2 are transcriptionally responsive to cI fusions.
Outline of Dual Bait System. (A) An activation domain-fused prey (P) interacts with a LexA-fused bait (B1) to drive transcription of lexAop-responsive LEU2 and lacZreporters, but does not interact with a cI-fused bait (B2) and, thus, does not turn on transcription of cIop-responsiveLYS2 and gusA reporters. The prey may represent a protein, as in Applications 1 and 3, or a peptide aptamer, as in Application 2. Note, in this example, the cI-Bait is drawn as representing a negative control for prey binding; the system can also be configured so that prey interacts with either or both baits, as inSerebriiskii et al. (1999). Points addressed in optimization of the Dual Bait two-hybrid system in this study (Table 1) are as follows: (1), varying expression level of baits; (2), enriching polylinkers to facilitate cloning of baits; (3), varying sensitivity of reporters; (4), diversifying plasmid antibiotic markers to facilitate isolation of library plasmid in E. coli. In addition, we have developed a robust yeast strain, SKY473, which is suitable both for bait testing and interaction mating in this system. (B) As described in Application 3, preys isolated against LexA-B1 and counterselected against cI-B2 are subsequently challenged with cI-B1 and LexA-B2, in a bait swap experiment. Those preys binding specifically to the B1 domain (as opposed to B1 specifically in the context of a LexA-B1 fusion protein) are retained preferentially. (C) One protein (P) may use different surface motifs to bind two different partners (B1, B2). (D) Application of a small molecule (drug or peptide) that obstructs one of the interactions shown in C will selectively turn off two of the four reporter genes, allowing subtractive scoring.
On the basis of prior experience optimizing and characterizing functionality of the Interaction Trap (Gyuris et al. 1993; Estojak et al. 1995), a simple two-hybrid precursor of the Dual Bait system, we have noted several points at which it is desirable to have reagent flexibility, to maximize the range of proteins that can be productively studied. These include control of bait expression level, inducible expression of baits, and range of reporter sensitivity. In addition, convenience of use is improved by the availability of bait expression plasmid variants with diverse polylinkers, plasmids with alternativeEscherichia coli selectable markers to aid plasmid recovery from yeast, robust strains useful for interaction mating approaches (Finley and Brent 1994), and antibodies allowing bait detection. We have developed an array of reagents that address these points for the Dual Bait. New reagents are described in Table1, whereas a complete set of available Dual Bait and compatible Interaction Trap reagents are summarized in Supplemental Table 1. Control data demonstrating the efficacy of the new reagents at achieving graduated bait expression and reporter induction are provided in Supplemental Figure 1. This information and detailed protocols for system use are also presented at our Web site (http://www.fccc.edu/research/labs/golemis/InteractionTrapInWork.html).
Summary of New Dual Bait-Compatible Reagents
We have now applied the enhanced Dual Bait reagents to a diverse group of biological problems involving determination of protein–protein interaction specificity. The results of four different categories of application are summarized below, with representative data.
Application 1
Identification of Proteins that Target a Specific Sequence Motif in a Larger Protein
In the first example, the Dual Bait system was used to identify and analyze effector proteins for members of the Rho subfamily of the Ras superfamily, namely Cdc42 and Rac. Activated Rac and Cdc42 induce actin reorganization and can themselves induce DNA synthesis and cell-cycle progression, and are required for transformation by Ras. The insert region found in Rho GTPases is a short (13 amino acid) sequence motif that is present in all Rho-family GTPases but absent from Ras and other small GTPases (Thapar et al. 2002). Deletion of the insert region from Rac1 or Cdc42 leads to their inability to transform cells, but not to the loss of other functions. These results imply that the insert region is required for the binding of one or more effectors that mediate the effects of Rac1 and Cdc42 on cell transformation.
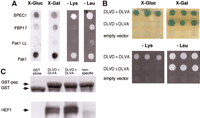
We adapted the Dual-Bait interaction trap system to find effectors specific for interaction with the insert region. LexA–Cdc42 L28 (encoding an activated, transforming allele of Cdc42) and cI-Cdc42 L28-Δ8 [activated Cdc42, insert region replaced with a sequence from Ras (Wu et al. 1998)] were used as selective baits to screen an AD-fused HeLa library. Among the isolated clones, two of note include SPEC1 (small protein effector ofCdc42; Pirone et al. 2000), which bound equivalently to Cdc42 with or without the insert region, and FBP17 (formin binding protein 17; Fuchs et al. 2001), which bound preferentially to Cdc42 containing the insert region (Fig. 2A). SPEC1 has been described previously as a Cdc42 interactor (Pirone et al. 2000), whereas FBP17 has been noted as having putative Rho-family interaction motifs (Fuchs et al. 2001), although no interaction has been demonstrated previously. Both proteins may serve as small GTPase effectors in control of the actin cytoskeleton, with further characterization of FBP17 in this context of particular interest, due to its interaction requirement for the insert region, and its recently determined role as a component of a translocation breakpoint in acute myelogenous leukemia (Fuchs et al. 2001).
Targeting of interacting proteins or peptides to small sequence motifs. (A) Differential interaction of Cdc42 effectors as detected by the Dual Bait procedure. Two Cdc42-interacting proteins, FBP17 (Fuchs et al. 2001) and SPEC1 (Pirone et al. 2000) were isolated by a yeast two-hybrid screen with LexA-Cdc42-L28. In this screen, 107diploids arising from an interaction mating (Finley and Brent 1994)-based screen yielded 25 positive colonies showing interactions with LexA-Cdc42 L28. Counterscreening against cI fused to activated Cdc42 lacking the insert region (Cdc42 L28-Δ8) revealed two clones, both containing the same cDNA, showing differential binding. All yeast shown contain both baits, and AD-fusions as indicated at left. Beside AD-fused FBP17 (specific for Cdc42-L28) and SPEC1 (bound both forms equivalently), AD-Pak1, which binds both forms of Cdc42, and AD-Pak1 LL, which cannot bind either form of Cdc42 (Sells et al. 1997), are included as controls. Results with growth and colorimetric reporters for Cdc42-L28-Δ8 [gusA (X-Gluc) and LYS2, first and third lines] and LexA-Cdc42-L28 [lacZ (X-Gal) andLEU2, second and fourth lines], are shown. (B) Selection of peptides specifically targeting HEF1-DLVD. A LexA-HEF1(DLVD) bait was used to identify interacting peptide aptamers, in collaboration with R. Finley. Selected peptides were rescreened in parallel against LexA-HEF1(DVLD) or cI-HEF1(DLVA), differing from the original bait by only a D363A substitution. Shown are peptides that interact with both DLVD and DLVA variants (DLVD = DLVA; activates all four reporters; representative sequence HHASTPRRESPGIMSPL); or with DLVD but not DLVA (DLVD>>DLVA; activates lacZ andLEU2 only; representative sequence, SGKFGEALPGWLSSACWCFG), and a nonspecific control peptide (negative; activates no reporters; representative sequence, EQLKYNRFWPWQWWGGRRLR). (C) Confirmation that peptides interact with HEF1 in an in vitrosystem, using pulldowns of endogenous HEF1 from cell lysates with the three GST-fused peptides from B, or with GST only. (Top) Levels of GST peptide or GST only in reaction; (bottom) associated HEF1.
Application 2
Identification of Peptides that Target a Specific Motif in a Larger Protein
HEF1 belongs to a family of focal adhesion-localized docking proteins that assemble protein complexes regulating cellular attachment, motility, apoptosis, and oncogenic transformation (O'Neill et al. 2000). As cells enter mitosis, the full-length HEF1 protein is cleaved at a DLVD consensus site for caspases 3 and 7, and the carboxy-terminal processed species is proteasomally degraded, leading to replacement of full-length HEF1 with an amino-terminal p55 species that localizes to the mitotic spindle (Law et al. 1998). Production of p55 is completely eliminated by the mutation of DLVD site to DLVA (Law et al. 1998). As a first step to studying potential mitosis-specific functions of HEF1, we wished to identify peptide aptamers (Colas et al. 1996) targeted to the DLVD motif that might regulate HEF1 cleavage.
A LexA-HEF1(DLVD) bait was used for interaction mating with a peptide aptamer library (in collaboration with R. Finley), and positive interactors rescreened against LexA-HEF1(DLVD) and cI-HEF1(DLVA). Of a series of peptides thus identified, some interacted with both DLVD and DLVA HEF1 variants (DLVD = DLVA), whereas some were specific to the DLVD WT form (DLVD>>DLVA), and hence, predicted to bind near the caspase consensus site (Fig. 2B). Binding of all peptides to endogenous HEF1 in cells was confirmed by pulldown with GST-fused peptides (DLVD = DLVA, DLVD > DLVA, or nonspecific) or GST only (Fig.2C). However, only peptides that bound DLVD, but not DLVA variants were able to selectively bind a DLVD, but not a DLVA mutant of full-length HEF1 in mammalian cells (E. Pugacheva, I. Serebriiskii, R. Finley and E. Golemis, unpubl.).
Application 3
Rapid Confirmation of Specific Interactions from a Library Screen
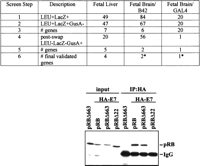
Some proteins of considerable biological interest, such as retinoblastoma (Rb), have a tendency to sometimes identify many nonspecifically interacting proteins from two-hybrid library screens, making it experimentally desirable to improve means to rapidly sort through sometimes large numbers of putative positives. In introducing this application, we note that alterations in the DNA sequence of retinoblastoma (Rb) gene contribute to the development of retinoblastoma with different penetrance. Low penetrant mutants give rise to retinoblastoma at a much lower frequency than expected for a null allele. Such mutants are expected to retain some protein-binding activities of wild-type pRB, which are of considerable interest for cancer biology, whereas high penetrant mutants are thought to have lost many of these interactions. RBΔ663 is a synthetic mutant that behaves similarly to the low-penetrant pRB mutants (see discussion in Sellers et al. 1998). The Dual Bait system was used to screen for proteins that interact with LexA-RBΔ663 but not with a high-penetrant (cI-RB Δex22, lacking exon 22) mutant of pRB (Fig. 3, top). In screening of three cDNA libraries, we identified several proteins that showed such binding specificity. Of interest, the E7 oncoprotein of human papilloma virus HPV-18 was isolated from two different libraries by this approach. E7, a known wild-type pRB interactor (Munger et al. 1989), was, for the first time, shown to interact with the low pentetrant RBΔ663 but not the high-penetrant Δex22 mutant.
Use of Dual Bait reagents to reduce false positive background. (Top) The RBΔ663 and RBΔex22 mutants of pRB are described in Sellers et al. (1998), and were expressed in the context of a large pocket domain of pRB containing amino acids from 379 to 928. SKY48 yeast expressing LexA-RBΔ663 and cI-RBΔex22 baits were used to screen three different libraries. (Lines 1,2) Numbers of clones positive for LexA-RBΔ663-responsive (LEU2,lacZ) but negative for cI-RBΔex22-responsive (gusA) reporters. (Line 3) Number of discrete genes represented among the clones. (Line 4) Number of clones positive for cI-RBΔ663-responsive (GusA), but not LexA-RBΔex22-responsive (LEU, lacZ) reporters; note, this reduction from line 2 values was not observed with retransformation testing with the original LexA-RBΔ663 and cI-RBΔex22 baits. (Line 5) Number of genes represented in line 4 set of clones. (Line 6) Number of surviving genes that were validated by coimmunoprecipiation and other techniques. (*) Note, papillomavirus E7 was identified from two different fetal brain libraries with B42 or GAL4 as activation domains. Although a legitimate pRB interactor, as E7 is not normally expressed in brain, it may represent an artifact of the libraries' construction. (Bottom) HA-tagged E7 and pRB derivatives were overexpressed in Saos-2 (Rb−/−) osteosarcoma cells and their interaction was determined by immunoprecipitation with anti-HA antibody (HA 11, BAbCO). The precipitated proteins pRB, pRBΔ663, and RBΔex22 (RBΔ22 in figure) were detected by immunoblotting with anti-RB antibody (XZ56). Input proteins are 10% of that used in immunoprecipitation.
As in Application 1, this is a demonstration of the capacity of the system to identify interactors with specific requirement for a binding motif in a larger protein. However, whereas in some libraries, positives represented multiple hits on a small number of genes, in others (fetal brain, GAL4), unique isolates of a large number of genes were obtained, suggesting specific interactions (Serebriiskii and Golemis 2001). To further improve specificity of the screen, we performed a bait swap, now expressing LexA-RBΔex22 and cI-RBΔ663 rather than LexA-RBΔ663 and cI-RBΔex22. Simultaneous retransformation of initially isolated preys with yeast containing swapped DBD fusions in parallel with the original baits eliminated a substantial number of the originally isolated clones, which were presumptively binding to a unique but artifactual configuration of the original baits (Fig. 3, top).
For the fetal brain/GAL4 library, this single step reduced the number of possible specific interacting clones from 20 to 1. With a single exception, all of the clones that interacted selectively with both LexA- and cI-RBΔ663 were confirmed by additional assays including coimmunoprecipitation (Fig. 3, bottom). The ease of swapping the two baits while remaining in the same reporter strain background, and using the same precalibrated reporter genes, is not matched by any other two hybrid-based system.
Application 4
Scoring Specific Disruption of Protein Interactions by Small Molecule Inhibitors
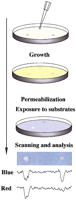
It has been of interest to try to develop small molecule inhibitors of specific protein–protein interactions as an intelligent way to manipulate signal transduction networks (discussed in Golemis et al. 2002). By use of a two-hybrid approach, small molecule inhibitors of the interaction between the oncoproteins Ras and Raf have been obtained (J. Kato-Stankiewicz, V. Khazak, I. Serebriiskii, E. Golemis, , in prep.). Using this compound as a tool in comparison with a general fungicide, we demonstrate that the Dual Bait reagents can be used in a single-step, plate-based colorimetric counterscreen that has the potential to facilitate future yeast-based screening for drugs that inhibit specific protein–protein interactions. As shown in Figure 4, yeast containing cI- and LexA- baits, and interacting preys, can be embedded as a lawn in soft agar medium, and subsequently overlaid with Magenta-Gal and X-Gluc. Interaction of Ras with its effectors Raf and RalGDS induces bothlacZ and gusA reporters, turning medium a uniform purple-blue, and thus providing a subtractive background to judge specific versus nonspecific reporter inhibition. Hence, whereas spotting a fungicide results in loss of both blue and red coloration, resulting in a white patch, and spotting of solvent has no effect, specific inhibition of the Ras–Raf interaction results in selective inhibition of GusA activation, resulting in a red patch. Prior discussions of drug screening applications of two hybrid have emphasized the use of counterselectable reporters such asURA3 (with 5-FOA) to score inhibition of interaction (Vidal and Endoh 1999); however, a difficulty with the use of counterselections in yeast is the frequency at which drug-insensitive mutants are likely to be selected. In this case, no growth pressure is applied, but selective differences in activity of a specific versus nonspecific reporter can be seen on the basis of color difference. Further, scanning of plates, and image analysis for red-blue ratio can provide quantitation of data; and given that pharmaceutical screens are standardly done using compounds arrayed in microtiter plates, automation of such an approach, potentially through adaptation of imaging technologies currently used for, for instance, microarrays, would be highly feasible.
Drug disruption of protein–protein interactions. (Top) Yeast containing baits and preys are mixed with low-melt agarose and poured over appropriate dropout growth medium. After agarose is set, 1 μL of each compound to be tested or solvent negative control is dropped on the plate. Yeast are incubated for 1–2 d, then permeabilized and overlaid with Z-buffer, Magenta-Gal (a red colorimetric substrate for LacZ) and X-Gluc (a blue colorimetric substrate for GusA). (Bottom) The result shown derives from a mixed population of yeast strains containing LexA-Ras and AD-Raf or cI-Ras and AD-RalGDS. In this example, only the colorimetric (lacZ andgusA) reporters are being assessed. Fungicide (left) inhibits both lacZ and gusA signal, whereas a specific Ras–Raf interaction inhibitor reduces only gusA(blue) output, leaving a red spot; solvent control produced no spots (data not shown). Shown below the spots are results obtained following a scan of plate, import of image into NIH Image, and performance of densitometry for signal intensity in blue versus red across the spot midline.
Summary
The development of a selective two-hybrid screening system has been of considerable general interest, and a number of groups have proposed strategies and reagent sets toward this end (discussed in Serebriiskii et al. 2001). For all new technologies, a key question is whether novel reagents demonstrated in proof-of-concept will prove broadly applicable, or problematic. We demonstrate that the enhanced Dual Bait reagents described here can be used in diverse applications involving proteins, peptides, and drugs, and in so doing, describe novel approaches that offer many advantages in probing and manipulating protein interaction specificity. We hope they will find productive use in the scientific community.
METHODS
Molecular and Microbiological Manipulation
Cloning of novel constructs was performed using conventional protocols. Yeast were cultured and manipulated by use of standard techniques. Details of the sequences and cloning sites encompassed in the panel of bait expression plasmids and reporter plasmids described in the Results section are available at (http://www.fccc.edu/research/labs/golemis/InteractionTrapInWork.html) and in Supplemental Figures 2–6. Further information concerning cloning strategies used for plasmid construction, or details of yeast strain construction, are available upon request, as are the plasmids and strains described herein. Expression of bait and prey proteins was confimed by Western analysis, with primary antibody to LexA, cI, or hemagglutinin (for preys).
For data presented in this work, two-hybrid experiments were performed as described (Estojak et al. 1995). For plate-based X-Gal and X-Gluc assays, chloroform/agarose overlay was used. For liquid β-glucuronidase or β-galactosidase assays, the procedure described in Serebriiskii et al. (1999) was used. Analysis of activation ofLYS2 or LEU2 reporters was accomplished by replica plating yeast to plates lacking leucine or lysine, and monitoring growth over 4 d.
WEB SITE REFERENCE
http://www.fccc.edu/research/labs/golemis/InteractionTrapInWork.html; Web site for this project.
Acknowledgments
These studies were supported by NIH grants GM54168 (to J.C.), CA63366 (to E.G.), and core grant CA06927 (to Fox Chase Cancer Center); and awards (to E.G.) from Ovarian SPORE and NCI Translational Pilot Projects, and the Pennsylvania Tobacco Health Research Formula Fund. E.N.P. is supported by DAMD 17-00-1-0429, from the Department of Defense Breast Cancer Research Program, managed by the U.S. Army Medical Research and Materiel Command. E.G. is a consultant to Morphochem. We thank Antje Taliana, Marijane Russell, and Michelle Berman for contributing work in the area of Dual Bait reagent construction. We gratefully acknowledge Russ Finley (Wayne State University) as a collaborator on dual bait peptide screening.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵6 Corresponding author.
-
E-MAIL EA_Golemis{at}fccc.edu; FAX (215) 728-3616.
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.450702.
-
- Received May 22, 2002.
- Accepted August 30, 2002.
- Cold Spring Harbor Laboratory Press