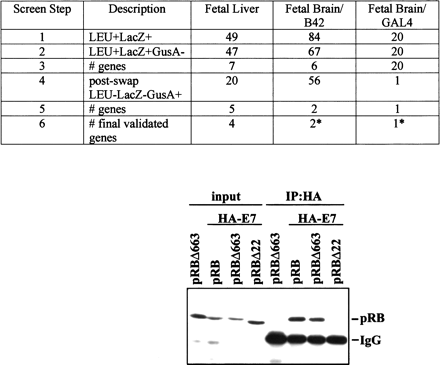
Use of Dual Bait reagents to reduce false positive background. (Top) The RBΔ663 and RBΔex22 mutants of pRB are described in Sellers et al. (1998), and were expressed in the context of a large pocket domain of pRB containing amino acids from 379 to 928. SKY48 yeast expressing LexA-RBΔ663 and cI-RBΔex22 baits were used to screen three different libraries. (Lines 1,2) Numbers of clones positive for LexA-RBΔ663-responsive (LEU2,lacZ) but negative for cI-RBΔex22-responsive (gusA) reporters. (Line 3) Number of discrete genes represented among the clones. (Line 4) Number of clones positive for cI-RBΔ663-responsive (GusA), but not LexA-RBΔex22-responsive (LEU, lacZ) reporters; note, this reduction from line 2 values was not observed with retransformation testing with the original LexA-RBΔ663 and cI-RBΔex22 baits. (Line 5) Number of genes represented in line 4 set of clones. (Line 6) Number of surviving genes that were validated by coimmunoprecipiation and other techniques. (*) Note, papillomavirus E7 was identified from two different fetal brain libraries with B42 or GAL4 as activation domains. Although a legitimate pRB interactor, as E7 is not normally expressed in brain, it may represent an artifact of the libraries' construction. (Bottom) HA-tagged E7 and pRB derivatives were overexpressed in Saos-2 (Rb−/−) osteosarcoma cells and their interaction was determined by immunoprecipitation with anti-HA antibody (HA 11, BAbCO). The precipitated proteins pRB, pRBΔ663, and RBΔex22 (RBΔ22 in figure) were detected by immunoblotting with anti-RB antibody (XZ56). Input proteins are 10% of that used in immunoprecipitation.











