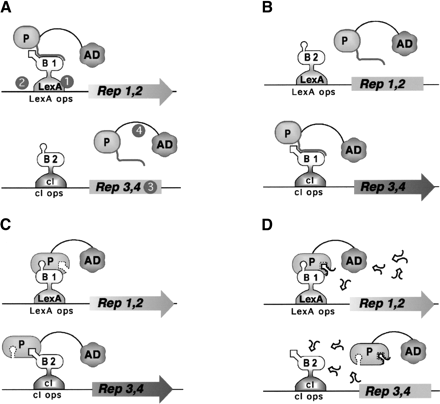
Outline of Dual Bait System. (A) An activation domain-fused prey (P) interacts with a LexA-fused bait (B1) to drive transcription of lexAop-responsive LEU2 and lacZreporters, but does not interact with a cI-fused bait (B2) and, thus, does not turn on transcription of cIop-responsiveLYS2 and gusA reporters. The prey may represent a protein, as in Applications 1 and 3, or a peptide aptamer, as in Application 2. Note, in this example, the cI-Bait is drawn as representing a negative control for prey binding; the system can also be configured so that prey interacts with either or both baits, as inSerebriiskii et al. (1999). Points addressed in optimization of the Dual Bait two-hybrid system in this study (Table 1) are as follows: (1), varying expression level of baits; (2), enriching polylinkers to facilitate cloning of baits; (3), varying sensitivity of reporters; (4), diversifying plasmid antibiotic markers to facilitate isolation of library plasmid in E. coli. In addition, we have developed a robust yeast strain, SKY473, which is suitable both for bait testing and interaction mating in this system. (B) As described in Application 3, preys isolated against LexA-B1 and counterselected against cI-B2 are subsequently challenged with cI-B1 and LexA-B2, in a bait swap experiment. Those preys binding specifically to the B1 domain (as opposed to B1 specifically in the context of a LexA-B1 fusion protein) are retained preferentially. (C) One protein (P) may use different surface motifs to bind two different partners (B1, B2). (D) Application of a small molecule (drug or peptide) that obstructs one of the interactions shown in C will selectively turn off two of the four reporter genes, allowing subtractive scoring.











