Isolation of Zebrafish gdf7 and Comparative Genetic Mapping of Genes Belonging to the Growth/Differentiation Factor 5, 6, 7 Subgroup of the TGF-β Superfamily
- Alan J. Davidson1,
- John H. Postlethwait2,
- Yi-Lin Yan2,
- David R. Beier3,
- Cherie van Doren3,
- Dorothee Foernzler3,
- Anthony J. Celeste4,
- Kathryn E. Crosier1, and
- Philip S. Crosier1,5
- 1Department of Molecular Medicine, School of Medicine, University of Auckland, Auckland, New Zealand; 2Institute of Neuroscience, University of Oregon, Eugene, Oregon 97403-1254 USA; 3Brigham and Women’s Hospital Harvard Medical School, Boston, Massachusetts 02115 USA; 4Genetics Institute, Cambridge, Massachusetts 02140 USA
Abstract
The Growth/differentiation factor (Gdf)5, 6, 7 genes form a closely related subgroup belonging to the TGF-β superfamily. In zebrafish, there are three genes that belong to the Gdf5, 6, 7subgroup that have been named radar, dynamo, andcontact. The genes radar and dynamo both encode proteins most similar to mouse GDF6. The orthologous identity of these genes on the basis of amino acid similarities has not been clear. We have identified gdf7, a fourth zebrafish gene belonging to the Gdf5, 6, 7 subgroup. To assign correct orthologies and to investigate the evolutionary relationships of the human, mouse, and zebrafish Gdf5, 6, 7subgroup, we have compared genetic map positions of the zebrafish and mammalian genes. We have mapped zebrafish gdf7 to linkage group (LG) 17, contact to LG9, GDF6 to human chromosome (Hsa) 8 and GDF7 to Hsa2p. The radar anddynamo genes have been localized previously to LG16 and LG19, respectively. A comparison of syntenies shared among human, mouse, and zebrafish genomes indicates that gdf7 is the ortholog of mammalian GDF7/Gdf7. LG16 shares syntenic relationships with mouse chromosome (Mmu) 4, including Gdf6. Portions of LG16 and LG19 appear to be duplicate chromosomes, thus suggesting thatradar and dynamo are both orthologs of Gdf6. Finally, the mapping data is consistent with contact being the zebrafish ortholog of mammalian GDF5/Gdf5.
[The sequence data described in this paper have been submitted to the GenBank data library under accession numbers AF113022 and AF113023.]
The TGF-β superfamily is a large group of genes encoding secreted signaling molecules that regulate a diverse range of biological processes during growth, repair, and embryonic development (Kingsley 1994; Hogan 1996). TGF-β-related peptides are synthesized as large precursor molecules comprised of these two major domains: a poorly conserved amino-terminal pre-prodomain and a highly conserved carboxy-terminal mature domain. For most superfamily members, the mature domain contains seven invariant cysteine residues that are involved in intramolecular and intermolecular disulphide bonds (Daopin et al. 1992; Schlunegger and Grütter 1992; Griffith et al. 1996; Eigenbrot and Gerber 1997). The active signaling molecule is a homo- or heterodimer of the mature domain that is released from the prodomain by cleavage at a dibasic R-X-X-R site (in which X is any amino acid; Dubois et al. 1995;Nachtigal and Ingraham 1996).
Members of the TGF-β superfamily can be organized into related subgroups on the basis of amino acid similarity within the mature domain. The mouse Growth/differentiation factor (Gdf)5, 6, and 7 genes were originally isolated from genomic DNA by a degenerate polymerase chain reaction approach and homologous genes have been identified in other mammals and zebrafish (Chang et al. 1994; Storm et al. 1994; Rissi et al. 1995; Bruneau and Rosa 1997; Bruneau et al. 1997; Wolfman et al. 1997). In the mouse and human, these genes show distinct patterns of expression in developing cartilage and joints (Chang et al. 1994; Storm et al. 1994; Storm and Kingsley 1996; Wolfman et al. 1997). Gdf5 was mapped to a region of mouse chromosome 2 that contains the brachypodismmutation. Mutations in Gdf5 were found to be responsible for the brachypodism phenotype (Storm et al. 1994), whereas mutations in the human ortholog (also known as CDMP1) cause the phenotypically similar human disorder Hunter-Thompson type chondrodysplasia (Thomas et al. 1996). In addition to their role in connective tissue development, GDF5, 6, and 7 also have effects on other tissues. A targeted mutation in the mouse Gdf7 gene results in hydrocephalus and a defect in the development of discrete dorsal commissural neurons (Lee et al. 1998). GDF6 and GDF7 inhibit terminal differentiation of myoblasts (Inada et al. 1996), whereas GDF5 can act as a neurotrophic and angiogenic factor (Krieglstein et al. 1995; Yamashita et al. 1997).
In zebrafish, three genes belonging to the Gdf5, 6,7 subgroup have been reported and named radar,dynamo, and contact (Rissi et al. 1995; Bruneau and Rosa 1997; Bruneau et al. 1997). Multiple tissues expressradar during embryonic development, including putative neural crest cells, the neural tube, and the retina (Rissi et al. 1995). Expression of dynamo is found in posterior neural tissue during the development of the central nervous system (Bruneau and Rosa 1997), whereas contact is expressed in the pectoral fin buds and the developing cartilage of the head (Bruneau et al. 1997). The orthologous assignment of radar, dynamo,contact, and their evolutionary relationship to mammalianGdf5, 6, 7 genes has been unclear.
In this report, we describe the isolation of zebrafish gdf7, a fourth gene belonging to the Gdf5, 6, 7 subgroup. Theradar and dynamo genes have been localized to LG16 and LG19 (Postlethwait et al. 1998). We have mapped genetically the remaining zebrafish genes gdf7 andcontact, as well as human GDF6 and GDF7. A comparison of syntenies shared among human, mouse, and zebrafish genomes suggests that gdf7 is the ortholog of mammalian GDF7/Gdf7. Both radar and dynamoappear to be orthologs of murine Gdf6 that have arisen from a chromosomal duplication event that has occured in teleosts. Finally, the mapping data is consistent with contact being the zebrafish ortholog of GDF5/Gdf5.
RESULTS
Isolation of gdf7 and Phylogenetic Analysis of the Gdf5, 6, 7 Subgroup
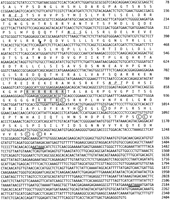
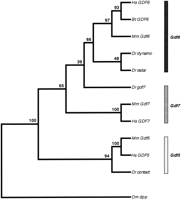
A genomic library was screened with a probe encoding the mature domain of murine GDF7. Twenty of the strongest positive plaques were further purified, subcloned, and sequenced. In addition to the previously published genes radar, dynamo, andcontact (Rissi et al. 1995; Bruneau and Rosa 1997; Bruneau et al. 1997), a fourth gene was identified that we have namedgdf7. Although a gdf7 cDNA failed to be isolated from two independent cDNA libraries, the ORF identified in the gdf7genomic fragment was extended by an additional 137 amino acids by the technique of 5′ rapid amplification of cDNA ends (5′ RACE) on cDNA derived from adult testis tissue. The combined nucleotide sequence and conceptual translation of gdf7 is shown in Figure1. An analysis of the gdf7 ORF failed to identify a potential start methionine and hydrophobic signal sequence. Repeated attempts of 5′ RACE were not successful in extending the coding sequence further. Therefore, the complete coding sequence forgdf7 remains to be isolated. Three consensus sites for N-linked glycosylation were identified in the prodomain followed by two potential processing sites that conform to the R-X-X-R motif. Proteolytic cleavage at these sites would predict mature GDF7 peptides of 130, 128, or 127 amino acids (site 1; boxed and shaded in Fig. 1) and 102 amino acids (site 2; boxed in Fig. 1), respectively. Two cleavage sites in similar positions are found within human GDF7 and would give rise to mature peptides of 129 or 104 amino acids (Wolfman et al. 1997). Similarly, murine GDF7 has two cleavage sites, although these are more distantly separated by a glycine rich insert (Storm et al. 1994). Both of the human forms have been expressed in E. coli and are reported to be biologically active (Wolfman et al. 1997). The mature domain encoded by gdf7, comprised of 113 conserved carboxy-terminal amino acids, was compared with the mature domains from mouse GDF5, GDF6, and GDF7 and found to share 81%, 88%, and 82% amino acid identities, respectively. Two other genes belonging to the zebrafish Gdf5, 6, 7 subgroup,radar and dynamo, also encode mature domains that are most similar to GDF6 with 92% and 88% amino acid identity, respectively (Rissi et al. 1995; Bruneau et al. 1997). On the basis of these sequence comparisons, the orthologous identities ofgdf7, radar, dynamo, and contactare not clear. Therefore, a neighbor-joining bootstrap tree was constructed to more accurately determine the evolutionary relationships between the zebrafish and mammalian genes (Fig. 2). On the basis of this analysis, contact groups with humanGDF5 and murine Gdf5 with high bootstrap support that is consistent with the suggestion that contact is the zebrafish ortholog of Gdf5 (Bruneau et al. 1997). Theradar and dynamo genes are most closely related to each other and are equally related to mammalian Gdf6orthologs. The orthologous identity of gdf7 cannot be unambiguously determined from the gene tree.
Nucleotide and predicted amino acid sequence of zebrafishgdf7. The ORF contains two potential proteolytic processing motifs (R-X-X-R); site 1 (boxed and shaded) and site 2 (boxed). Three N-linked glycosylation sites are located within the prodomain (underlined and in italics) and the mature domain contains seven conserved cysteine residues (circled). The sequence of the putative 3′ UTR contains two potential polyadenylation signals (thick underlined). (Arrowhead) Boundary between sequence obtained by 5′ RACE and sequence obtained from genomic DNA. The GenBank accession nos. for zebrafish gdf7 are AF113022 and AF113023.
Phylogenetic relationships among members of the zebrafish and mammalianGdf5, 6, 7 subgroup of genes. A neighbor-joining bootstrap tree was generated from mature domain sequences and arbitrarily outgrouped to dpp. Numbers above each branch point indicate percent bootstrap values (1000 replications). On the basis of this analysis and the genetic mapping (Fig. 3), the genes can be classified into three orthology groups:Gdf5, Gdf6, and Gdf7 as indicated by the three shaded bars. (Bt) Bos taurus; (Dr) Danio rerio; (Hs) Homo sapiens; (Mm) Mus musculus; (Dm)Drosophila melanogaster.
Genetic Mapping and Analysis of Shared Syntenies
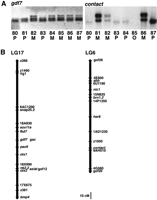
To map gdf7, we identified a restriction fragment length polymorphism (RFLP) in the putative 3′ untranslated region (UTR) and used this to localize the gene to linkage group (LG) 17 (Fig.3). LG17 shares syntenies with three mouse chromosomes (Mmu); Mmu14 (bmp4/Bmp4 and otx2/Otx2), Mmu2 (snap25.2/Snap25 andaxial/Hnf3β), and Mmu12 (gsc/Gsc,pax9/Pax9, nk2.2/Nkx2-1,fkd7/Hnf3α, and sox11a/Sox11; Fig.4A). Mmu12 also contains Gdf7. LG17 shares syntenies with three human chromosome (Hsa) arms; Hsa2p (otx1/OTX1 and sox11a/SOX11; S.T. De Martino, T. Jowett, Y. Yan, J. Postlethwait, A. Ashworth and C.A. Austin, unpubl.), Hsa20p (snap25.2/SNAP25 andaxial/HNF3β) and Hsa14q (bmp4/BMP4,gsc/GSC, pax9/PAX9, nk2.2/NKX2A, otx2/OTX2, andfkd7/HNF3α;Postlethwait et al. 1998; Fig. 4A,B). To map human GDF7, primers were designed to the human sequence (A.J. Celeste, unpubl.) and scored on the Stanford Human Radiation Hybrid G3 mapping panel.GDF7 mapped to the short arm of Hsa2 near marker SHGC-33991 (lod score 12.13, cR_10,000, 5.02 cR). SHGC-33991 is in the interval from 40.7 to 48.5 cm that places it at the cytological postion 2p22-p21. Therefore, the three zebrafish/human gene pairs gdf7/GDF7 (2p22-p21), sox11a/SOX11 (2p25), andotx1/OTX1 (2p13) form a conserved synteny comprising part of Hsa2p and the central portion of LG17. On the basis of the shared syntenies in the mammalian and zebrafish genomes, it is likely that the zebrafish gdf7 gene is the ortholog of GDF7/Gdf7.
Genetic mapping of gdf7 and contact. (A) A RFLP was identified for gdf7 by the restriction endonucleaseTaq1. SSCP analysis was used to detect a polymorphism in thecontact gene. Below each panel are the genotypes (M, maternal allele; P, paternal allele; O, missing data) of eight selected F2 progeny (numbers 80–87) from the mapping panel. (B) Genetic maps of LG17 and LG6 showing the location ofgdf7 and contact. Cloned genes are written in italics, microsatellite markers begin with z or gof and RAPD markers begin with a number (Postlethwait et al. 1994; Knapik et al. 1996,1998; Postlethwait et al. 1998).
Comparative genetic mapping of the Gdf5, 6,7 subgroup of genes in zebrafish and mammals. (A)Gdf7-bearing chromosome group. (B)Gdf5-bearing chromosome group. (C)Gdf6-bearing chromosome group. Each row shows the contents of different zebrafish (LG), human (Hsa), or mouse (Mmu) chromosomes. For ease of comparison, the genes have been arbitrarily ordered so that members of gene families are displayed in columns. Within a chromosome group the genes in columns are apparent orthologs, or in the case of some zebrafish chromosomes, paralogs resulting from a genome duplication not shared with tetrapods.
A polymorphism in contact was identifed by single-strand conformation polymorphism (SSCP) analysis and used to localize this gene to LG6 (Fig. 3), which also contains the genes brn1.2,ehh, and nic1 (Fig. 4B). LG6 and LG9 appear to be duplicate chromosomes (Amores et al. 1998; Postlethwait et al. 1998) and contain the genes brn1.1 and brn1.2, respectively, which are equally related to mouse Brn1 (Sampath and Stuart 1996). This suggests that the duplication event that generated LG6 and LG9 arose after the divergence of ray-finned (modern bony fish ancestor) and lobe-finned (tetrapod ancestor) fish ∼420 million years ago (Ahlberg and Milner 1994). LG9 contains a number of other loci that have mammalian orthologs on Hsa2q, Mmu1, and Mmu2 (Fig.4B). LG6 contains nic1, which encodes the nicotinic acetylcholine receptor-α (Sepich et al. 1998), and is the zebrafish ortholog of human CHRNA1 on Hsa2q and mouse Acra on Mmu2. Mmu2 also contains Gdf5, which is likely to be orthologous to contact, on the basis of the phylogenetic analysis (Fig. 2). Taken together, the mapping data suggests that an ancient chromosome may have contained the progenitors ofcontact/GDF5/Gdf5, bmp2b/BMP2/Bmp2, snap25.1/SNAP25/Snap25, PAX1/Pax1, HNF3β/Hnf3β, brn1.1/brn1.2/Brn1, hoxd4/HOXD4/Hoxd4, evx2/EVX2/Evx2, eng1/EN1/En1, dlx2/DLX2/Dlx2, des/DES/Des,ehh/hha/IHH/Ihh, actr2/Acvr2a, actbb/INHBB/Inhbb, dermo1/DERMO1, andnic1/CHRNA1/Acra. In the human lineage, this was broken into two segments found on Hsa20 and Hsa2q. In the mouse chromosome, after some inversions that altered the gene order with respect to humans, it was also broken into two chromosomes, but in a different place than in the human lineage, thereby giving Mmu1 and Mmu2. In the zebrafish lineage, the ancestral chromosome or at least a segment of it, was duplicated, eventually giving rise to portions of LG6 and LG9. The segment of LG20 that contains bmp2b and snap25.1probably became separated from either LG6 or LG9 by translocation.
The genes radar and dynamo have been localized to LG16 and LG19, respectively (Postlethwait et al. 1998). The phylogenic analysis (Fig. 2) suggests that radar and dynamo are most closely related to each other and are equally related to mammalianGDF6/Gdf6. Therefore, radar and dynamo may be duplicate genes that are orthologous to GDF6/Gdf6. The mapping data further supports this conclusion. LG16 containsradar, zp50pou, zp47pou, hoxa13a, and evx1 (Amores et al. 1998; Postlethwait et al. 1998; Fig.4C). The murine orthologs of zp50pou and zp47pou are found on Mmu4 (Avraham et al. 1993; Hauptmann and Gerster 1996; Spaniol et al. 1996), which also contains Gdf6. Therefore, these conserved syntenies are consistent with radar being orthologous to murine Gdf6. LG19 contains dynamo,hoxa5b, dlx6, and npy (Fig. 4C; Amores et al. 1998; Postlethwait et al. 1998). Gene phylogenies show that thehoxaa and hoxab loci in zebrafish are paralogs that appear to have been derived from a chromosomal duplication in the teleost lineage (Amores et al. 1998). The linkage of radar anddynamo to paralogous hox clusters is consistent with the suggestion that these zebrafish Gdf5, 6,7 subgroup members are duplicate orthologs of murineGdf6. To map human GDF6, primers were designed to the human sequence (A.J. Celeste, unpubl.) and scored on the Genebridge4 radiation hybrid panel. GDF6 mapped to Hsa8 near marker WI-9077 (lod score >15, cR_3000, 5.23 cR), which corresponds to the gene ATP6D (UniGene Hs.86905). WI-9077 maps in the interval from D8S270 (102.1 cm) to D8S257 (110.3 cm) on the human gene map that corresponds to the cytogenetic position 8q21.3. This region of Hsa8 shares syntenies with Mmu4 including the genes CBFA2/Cbfa2 and CALB1/Calb, consistent with the chromosomal location of Gdf6 in the mouse Fig. 4C). The human orthologs of zp50pou/Pou3f1 andzp47pou/Pou3f2 are found on 1p34.1 and 6q16, respectively (Sumiyama et al. 1998; Atanasoski et al. 1995), suggesting that these genes became dispersed from GDF6, CBFA2, andCALB1 by independent rearrangements in the human lineage.
In comparing Figure 4, A and B, notice that the genesGDF5/Gdf5, BMP2/Bmp2, PAX1/Pax1, andNkx2-2 on Hsa20 and Mmu2 are paralogous to GDF7/Gdf7,BMP4/Bmp4, PAX9/Pax9, and NKX2A/Nkx2-1 on Hsa2p or Hsa14 and Mmu12 or Mmu14. This suggests that an ancient duplication event that occurred prior to the divergence of the ray-finned and lobe-finned fishes produced two duplicate chromosome segments. Independent rearrangements then occurred in the murine and human lineages resulting in these segments becoming portions of Hsa20, Hsa14, Hsa2p, and Mmu2, Mmu12, and Mmu14.
Expression of gdf7
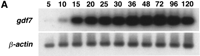
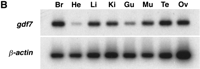
The expression of gdf7 during embryonic development and in adult tissues was examined by RT-PCR by use of gdf7-specific primers (Fig. 5). Developmental stages from 5 hr postfertilization (hpf) to 5 days postfertilization and the organs brain, heart, liver, kidney, gut, muscle, testis, and ovary were examined. The PCR products were analyzed by Southern hybridization with an internal radiolabeled oligonucleotide specific to gdf7. Expression of gdf7 was weakly detected at 5 hpf and then at every stage examined through to 120 hpf. Expression of gdf7was detected in all of the adult tissues examined but was most readily amplified from cDNA isolated from the brain, liver, kidney, muscle, testis, and ovary. No RT-PCR products were detected in control samples that were not treated with reverse transcriptase (data not shown). As a positive control, the same cDNAs were amplified withβ-actin-specific primers and also hybridized to an internal oligonucleotide.
Embryonic and adult expression of gdf7. Total RNA was isolated from zebrafish embryos at selected stages from 5 hpf to 120 hpf (A) and from adult tissues (B). The expression ofgdf7 was examined by RT-PCR and the amplified products were detected following hybridization of an internal radiolabeledgdf7-specific oligonucleotide. The expression ofβ-actin was examined as a positive control for cDNA synthesis. (Br) brain; (He) heart; (Li) liver; (Ki) kidney; (Gu) gut; (Mu) muscle; (Te) testis; (Ov) ovary.
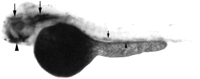
The expression of gdf7 at 48 hpf was examined by whole mount in situ hybridization (Fig. 6). Transcripts forgdf7 were detected in dorsal and ventral regions of the head, the pronephric ducts, and the dorsal aorta in the trunk of the embryo. In the head, the dorsal population of cells formed two bilateral stripes that appeared coincident with the location of the paired trabeculae of the developing neurocranium (Schilling et al. 1996;Schilling and Kimmel 1997). The more ventrally located cells that express gdf7 are likely to represent precartilaginous condensations of the palatoquadrates, ceratohyals, and Meckel’s cartilage, which comprise the jaw and supporting elements (Schilling et al. 1996; Schilling and Kimmel 1997). A more detailed examination ofgdf7 expression during craniofacial development and how it compares with other members of the zebrafish Gdf5, 6, 7 subgroup is currently under way.
Expression of gdf7 at 48 hpf by whole mount in situ hybridization. A lateral view of a 48 hpf embryo is shown with anterior toward the left and dorsal toward the top of the page. Transcripts forgdf7 can be detected in two bilateral stripes of cells in regions of the developing neurocranium (large arrows) and in precartilage condensations of the developing jaw (large arrowhead). Expression of gdf7 is also observed in the dorsal aorta (small arrow) and in the pronephric ducts (small arrowhead). The staining in the yolk is nonspecific.
DISCUSSION
We have identified gdf7, a fourth gene member of the zebrafish Gdf5, 6, 7 subgroup. The predicted amino acid sequence encoded by gdf7 contains potential processing sites and seven conserved cysteine residues characteristic of members of the TGF-β superfamily. An analysis of gene expression by RT-PCR showed that gdf7 is an active gene that is expressed during embryonic development and in adult tissues. Expression ofgdf7 in 48 hpf embryos was examined by whole mount in situ hybridization and transcripts were predominantly detected in regions of the head undergoing cartilage development.
The finding that the zebrafish genome contains four genes that belong to the Gdf5, 6, 7 subgroup prompted us to explore the evolutionary origin of these genes by comparative genetic mapping. Such an approach provides an alternative basis on which to infer evolutionary relationships among distantly related phyla. Thegdf7 gene mapped to LG17, which shares syntenic relationships with genes found on Mmu12, including Gdf7. Similarly, LG17 shares syntenies with Hsa2p that includes the GDF7 gene. Thus, it is likely that gdf7 is the zebrafish ortholog ofGDF7/Gdf7. Additional support for this orthologous assignment is the presence of two potential cleavage sites in the prodomain of zebrafish GDF7 that are also found conserved in human and mouse GDF7 sequences but are not found within human or mouse GDF5 or GDF6 (Storm et al. 1994; Wolfman et al. 1997). Furthermore, the partial prodomain sequence of zebrafish GDF7 shows the greatest similarity to the prodomain of human GDF7 (data not shown). The contact gene is most closely related to mammalian GDF5/Gdf5 genes and its chromosomal localization to LG6 is consistent with it being the zebrafish ortholog of GDF5/Gdf5. The mapping data suggests LG6 and LG9 are duplicate chromosomes and a translocation has separated the genes bmp2b and snap25.1 from either LG6 or LG9. Another ortholog of bmp2 exists in zebrafish, termedbmp2a (Martínez-Barberá et al. 1997) and maps to LG17 (J.H. Postlethwait, unpubl.), which also containssnap25.2. This data suggests that parts of LG17 and LG20 are duplicate chromosome segments and raises the possibility that anothercontact gene may also exist. If so, it would likely map to either LG9, LG17, or LG20.
It has been suggested that dynamo may represent a new member of the Gdf5, 6, 7 subgroup that has yet to be identified in higher vertebrates (Bruneau and Rosa 1997). However, our data suggests an alternative hypothesis. The radar anddynamo genes appear as sister groups on the gene tree and are closely related to mammalian GDF6/Gdf6 orthologs. Genetic mapping localized radar and dynamo to LG16 and LG19, respectively, and previous mapping data suggests that these are duplicate chromosomes as they each contain one copy of duplicatehoxa complexes (Amores et al. 1998). LG16 shares syntenic relationships with genes found on Mmu4, including Gdf6. Therefore, it is likely that dynamo and radar both represent orthologs of murine Gdf6 that have arisen from a chromosomal duplication that has occurred in the zebrafish lineage. Members of the Gdf5, 6, 7 subgroup share a similar gene structure comprised of two coding exons (A.J Celeste, unpubl.). The location of the intron separating the two coding exons is specific for each subgroup member. The position of the intron separating the two coding exons in radar and dynamois conserved, consistent with the suggestion that these are duplicated genes (A.J. Davidson, unpubl.). Thus, we would predict that an additional Gdf5, 6, 7 member corresponding to the ortholog of dynamo will not be found in mammals. Extensive genomic library screening has yet to reveal a dynamoortholog in humans (A.J. Celeste, unpubl.).
It is believed that gene duplication by polyploidization has been an important part of vertebrate evolution and a major contributor to multigene families (Ohno 1970; Holland et al. 1994; Sidow 1996;Postlethwait et al. 1998). At least two genome duplications have occurred during the evolution of vertebrates, both happening prior to the divergence of ray-finned and lobe-finned fish (Holland et al. 1994;Amores et al. 1998; Postlethwait et al. 1998). A comparison of shared syntenies in this study suggests that the loci gdf7/GDF7/Gdf7are paralogs of contact/GDF5/Gdf5 and probably arose from one of these rounds of vertebrate polyploidization. However, it is apparent from the mapping data that additional chromosome duplications have since occurred during zebrafish evolution, such as those that generated the LG6/LG9 and LG16/LG19 pairs of chromosome segments. Extra members of other developmental multigene families in zebrafish have also been reported (Akimenko et al. 1995; Ekker et al. 1995; Stock et al. 1996;Zardoya et al. 1996) and in some cases these extra genes are found in duplicated chromosome segments (Postlethwait et al. 1998). A comprehensive screen for zebrafish hox genes and the subsequent mapping of these genes has revealed that an additional genome duplication event not shared with tetrapods is likely to have occurred prior to teleost radiation (Amores et al. 1998). Such a tetraploidization event would explain the greater number of paralogous gene copies found in zebrafish and provides a mechanism for the origin of the gene duplicates radar and dynamo.
By determining the orthologous assignments of gdf7,radar, dynamo, and contact it will now be possible to interpret more meaningfully a comparison of expression patterns between zebrafish and mammals that should yield additional insights into the function of these genes during development. Furthermore, the mapping of these genes will aid in the identification of candidates for some of the mutations recently generated in zebrafish, particularly those involved in cartilage and bone formation. Finally, in line with agreed on nomenclature (Westerfield 1994) and to avoid further confusion resulting from orthologs having multiple names, we propose that radar, dynamo, and contactbe renamed gdf6a, gdf6b, and gdf5, respectively.
METHODS
Isolation of Zebrafish gdf7
A 291-bp PCR fragment encoding most of the mature domain of murine GDF7 (Wolfman et al. 1997) was radioactively labeled by random priming (Boehringer Mannheim Australia, NSW) and used as a probe to screen ∼1 × 106 plaques of a zebrafish genomic DNA library (Stratagene, La Jolla, CA). Hybridization was carried out according to standard procedures (Sambrook et al. 1989) at 65°C in 5× SSC, 5× Denhardt’s solution, 0.5% SDS and 100 μg/ml denatured salmon sperm DNA. The membranes (Hybond-N+; Amersham, Buckinghamshire, England) were washed to a final stringency of 2× SSC, 0.1% SDS at 65°C. Hybridizing plaques were picked and purified by two additional rounds of screening. Four distinct positive phage clones corresponding to gdf7, radar, dynamo, and contactwere identified and the sequences that hybridized to the probe were subcloned into pBluescript SK- (Stratagene, La Jolla, CA) and sequenced. Additional sequence of gdf7 was identified by use of total RNA extracted from adult testis tissue and the 5′ RACE system (GIBCO–BRL, Gaithersburg, MD) according to the manufacturer’s instructions.
DNA Sequencing
DNA sequencing of both strands was done with the PRISM Ready Reaction kits with AmpliTaq FS DNA Polymerase (Perkin Elmer, Foster City, CA), following the manufacturer’s recommendations for template and primer concentrations and cycling conditions. M13 forward and reverse primers or synthetic oligonucleotide primers were used. Reactions were run in the 9600 GeneAmp PCR system (Perkin Elmer, Foster City, CA) or the PTC200 Peltier Thermal Cycler (MJ Research, Watertown, MA). Sequencing reactions were purified by 96-well gel filtration blocks (AGCT Technologies, Gaithersburg, MD) and then resolved on an acrylamide gel with the Perkin Elmer 373 DNA analysis system. Sequence assembly and editing was performed on Apple Macintosh computers with the Sequencher program (Gene Codes, Ann Arbor, MI).
Phylogenetic Analysis
Amino acid sequences of the following proteins (GenBank accession nos. or the literature source are given in parentheses) were aligned by use of the software package MacVector (Oxford Molecular Group, Oxford, England): Radar (Rissi et al. 1995); zebrafish GDF7 (present study); Dynamo (X99769); Contact (Y12005); bovine GDF6 (also known as CDMP2;P55106); Decapentaplegic (U63857); murine GDF5, 6, 7 (U08337-U08839); human GDF5 (also known as CDMP1; P43026); and human GDF6 and GDF7 (Wolfman et al. 1997; additional unpublished sequence was provided by A.J Celeste). The alignment started 11 amino acids from the first conserved cysteine residue through to the last carboxy-terminal amino acid residue (113 characters). The phylogenetic analysis was performed with the Phylogenetic Inference Package, PHYLIP 3.5 (Felsenstein 1993). SEQBOOT was used to resample the alignment (1000 bootstrap replications). Distance matrices were generated by PROTDIST, on the basis of the Dayhoff PAM model of amino acid substitution. Neighbor-joining trees were calculated by NEIGHBOR and a consensus tree was derived by CONSENSE. Decapentaplegic (DPP) was used as an arbitrary outgroup for the tree.
Genetic Mapping
The gdf7 gene was mapped by the identification of a RFLP. Primers specific to the putative 3′ UTR of gdf7 were used to amplify a 221-bp fragment by use of genomic DNA from C32 or SJD parental strains as a template (a gift from L.I. Zon, Children’s Hospital, Boston, MA). The PCR reaction mixture contained ∼200 ng of genomic DNA, 1× PCR buffer (Perkin Elmer, Norwalk, CT), 1.5 mm MgCl2, 0.2 mm each dNTP, 2.5 units of AmpliTaq DNA Polymerase (Perkin Elmer, Norwalk, CT), and 1 μm of each primer (upper primer 5′-ATGACACCACATTTGGCTTGGG-3′; lower primer 5′-CACACCCTCTCAGTCAATGTAG-3′). Amplification was for 40 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C. The PCR products were subcloned into pCR2.1 (Invitrogen, San Diego, CA) and sequenced. A single base pair polymorphism was found that generated an additional Taq1 restriction enzyme site within the SJD amplified fragment.
For SSCP analysis, genomic DNA from C32 and SJD parental strains was amplified with primers specific to the 3′ UTR of contact(upper primer 5′-GTACGAGGACATGGTGGTGGAGAG-3′; lower primer 5′-TCGGAATGGAACTGAGTGAGAATG-3′). One of the primers was end-labeled by use of [γ-32P]ATP (6000 Ci/mmole; Amersham) and T4 polynucleotide kinase (GIBCO–BRL, Gaithersburg, MD). The PCR reaction mixture contained 250 ng of template DNA, 10 mm Tris-Cl (pH 8.3), 50 mm KCl, 2 mmMgCl2, 0.2 mm of each dNTP, 1 μm of each primer and 1.0 units of Taq DNA polymerase in a final volume of 12 μl. The reaction mixture was initially denatured at 94°C for 5 min. Amplification was for 30 cycles of 1 min at 94°C, 2 min at 55°C and 3 min at 72°C. There was a final extension of 7 min at 72°C. One-sixth of the sample was mixed with 8 μl of Stop solution (100% formamide, 0.25% Bromophenol Blue, 0.25% Xylene Cyanol FF), denatured for 5 min at 94°C and then chilled on ice before being loaded onto a 5% nondenaturing polyacrylamide gel containing 0.5× TBE buffer. The gel was electrophoresed at 4°C in 0.5× TBE buffer at 40 Watts. The gel was transferred and dried onto Whatman 3MM paper before being exposed to X-ray film at −70°C overnight.
The RFLP and SSCP identified for gdf7 and contactwere genotyped on DNA from 96 F2 progeny from the linkage map cross that has been genotyped previously for >650 PCR-based markers (Postlethwait et al. 1994; Johnson et al. 1996; Knapik et al. 1996,1998; Postlethwait et al. 1998). The strain distribution patterns were analyzed by the program LINKER (Postlethwait et al. 1994) and maps were constructed with MAPMAKER (Lander 1987). The locations of mammalian gene loci were taken from the Mouse Genome Database (http://www.informatics.jax.org/), the Online Mendelian Inheritance of Man (http://gdbwww.gdb.org/omim/docs/omimtop.html), the Genome Database (http://gdbwww.gdb.org/gdb), and The Human Transcript Map (http://www.ncbi.nlm.nih.gov/SCIENCE96/).
To map human GDF7, the primers 5′-GGATAGCCCGGGCGAAGACG-3′ and 5′-GCGGGGCCTCCTAACAGCAAATG-3′ were designed and found to amplify human, but not hamster, DNA from the Stanford Human Radiation Hybrid G3 Panel obtained from Research Genetics, Inc. DNAs from panel members were amplified by PCR and displayed on 2% agarose gels. Gels were scored and the results submitted to the Stanford Human Genome Center for mapping (http://www-shgc.stanford.edu/Mapping/rh/). To map human GDF6, two independent primer pairs (GDF6a: upper primer 5′-ACAAGCAGTACGAGGACATGG-3′, lower primer 5′-ATCCAGGCTGTTCCCTCAC-3′; GDF6b: upper primer 5′-AGAGAGGCGGAGGAGGAAG-3′, lower primer 5′-GGGCTGGCAGGTAGAAGTTAC-3′) were tested with the Genebridge4 radiation hybrid panel obtained from Research Genetics, Inc. Samples that gave concordant results with these primers were scored and the data analyzed by use of the Whitehead Institute radiation hybrid mapping web interface (http://carbon.wi.mit.edu:8000/cgi-bin/contig/rhmapper.pl).
RT–PCR and Whole Mount in Situ Hybridization
Total cellular RNA was isolated from embryos at 5, 10, 15, 20, 25, 30, 36, 48, 72, 96, and 120 hpf and from adult tissues with TRIZOL (GIBCO–BRL, Gaithersburg, MD) according to the manufacturer’s instructions. First strand cDNA was synthesized from each stage with Superscript II reverse transcriptase (GIBCO–BRL, Gaithersburg, MD) in a reaction volume of 20 μl according to the manufacturer’s instructions. To control for genomic DNA contamination, the samples were also incubated in the absence of reverse transcriptase. PCR was performed in a reaction volume of 50 μl containing 3 μl of cDNA, 1× PCR buffer (Perkin Elmer), 1.5 mmMgCl2, 0.2 mm of each dNTP, 2.5 units of AmpliTaq DNA Polymerase (Perkin Elmer, Norwalk, CT) and 1 μm of each gdf7-specific primer (upper primer 5′-TGGAAGACGGAGGACACG-3′; lower primer 5′-CTACCTGCACCCACAACT-3′) orβ-actin-specific primer (upper primer 5′-GTCGTCGACAACGGCTCCGGCATGTG-3′; lower primer 5′-CATTGTAGAAGGTGTGGTGCCAGAT-3′). Amplification was for 28 cycles of 1 min at 94°C, 1 min at 57°C and 1.5 min at 72°C generating a 328-bp gdf7 product or a 253-bpβ-actin product. The PCR products were resolved on a 1.5% agarose gel, blotted to nylon membranes (Hybond-N+; Amersham), and hybridized to an end-labeled internal oligonucleotide (gdf7-specific 5′-CAGAGTCCACCCCTCCC-3′;β-actin-specific 5′-GGACAGAAAGACAGCTACGT-3′) according to established procedures (Sambrook et al. 1989).
Whole-mount in situ hybridization was performed as described (Schulte-Merker et al. 1992). The gdf7 riboprobe was labeled with digoxigenin and detection of the antidigoxigenin antibody-alkaline phosphatase conjugate was done with 4-nitroblue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3-indolyl-phosphate (BCIP). After whole-mount in situ hybridization, the embryos were refixed in 4% paraformaldehyde, transferred into 80% glycerol, and photographed.
Acknowledgments
We thank the following people: Maria Vitas and Scott Mead for technical assistance; Kevin Bean for DNA sequencing; Dr Hazel Sive, Dr Leonard Zon, Steve Clark, Rod Hewick, Vicki Rosen, Jenn Dube, and Beth Murray for advice and assistance. This work was supported by a University of Auckland Doctoral Scholarship (A.J.D.), an Auckland Medical Research Foundation Senior Scholarship (A.J.D) and grants R01RR10715 (J.H.P) and PHS P01HD22486 (J.H.P).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵5 Corresponding author.
-
E-MAIL ps.crosier{at}auckland.ac.nz; FAX (649) 373-7492.
-
- Received August 26, 1998.
- Accepted December 7, 1998.
- Cold Spring Harbor Laboratory Press