Identification of Target Sites of the α2–Mcm1 Repressor Complex in the Yeast Genome
Abstract
The α2 and Mcm1 proteins bind DNA as a heterotetramer to repress transcription of cell-type-specific genes in the yeastSaccharomyces cerevisiae. Based on the DNA sequence requirements for binding by the α2–Mcm1 complex, we have searched the yeast genome for all potential α2–Mcm1 binding sites. Genes adjacent to the sites were examined for expression in the different cell mating types. These sites were further analyzed by cloning the sequences into a heterologous promoter and assaying for α2–Mcm1-dependent repression in vivo and DNA-binding affinity in vitro. Fifty-nine potential binding sites were identified in the search. Thirty-seven sites are located within or downstream of coding region of the gene. None of the sites assayed from this group are functional repressor sites in vivo or bound by the α2–Mcm1 complex in vitro. Among the remaining 22 sites, six are in the promoters of known α-specific genes and two other sites have an α2–Mcm1-dependent role in determining the direction of mating type switching. Among the remaining sequences, we have identified a functional site located in the promoter region of a previously uncharacterized gene, SCYJL170C. This site functions to repress transcription of a heterologous promoter and the α2–Mcm1 complex binds to the site in vitro. SCYJL170C is repressed by α2–Mcm1 in vivo and therefore using this method we have identified a new a-specific gene, which we callASG7.
Many eukaryotic transcription factors have been identified based upon their strong amino acid sequence conservation to known DNA-binding domains and regulatory proteins. Often, subsequent analysis of the temporal or spatial expression of these proteins suggests that they may be involved in the regulation of critical cellular or developmental processes. To understand their roles in developmental and transcriptional regulation, the identification of their target genes is essential.
One common method used to identify the target genes is to remove or alter the activity of the regulatory protein, either through mutations or by changing its expression, and then examine the effects of these changes. The downstream target genes can be identified through subtractive DNA libraries, differential display, or by DNA microarray techniques (DeRisi et al. 1996; Lashkari et al. 1997; Chu et al. 1998;Lutfiyya et al. 1998; Roth et al. 1998). This approach works well for organisms such as the yeast Saccharomyces cerevisiae, in which it is possible to construct strains with a mutation in these genes and assay the effects of mutants (DeRisi et al. 1997; Lutfiyya et al. 1998). In higher eukaryotes, however, it is difficult or impossible to use this approach to screen for downstream target genes because it is difficult to alter the activity of the regulatory proteins.
As an alternative approach to identifying the target genes of transcriptional regulatory proteins, the binding sites for these proteins can be determined through in vitro site selection methods (Blackwell and Weintraub 1990). This information is then used to search the genomic databases for genes containing these sites. The lack of a complete sequence of the organism has made this approach limited in the past. The genomic DNA sequences of many prokaryotic organisms, as well as the eukaryotes S. cerevisiae and Caenorhabditis elegans, however, have been reported over the last few years (Fleischmann et al. 1995; Bult et al. 1996; Goffeau et al. 1996;Deckert et al. 1998). The genome sequence projects of several other model organisms, such as Drosophila and Arabidopsis, are well under way (Dickson 1998; Flanders et al. 1998; Venter et al. 1998). As progress on the genome projects continues, it becomes possible to search an entire genome for all potential binding sites of a given DNA-binding protein. In this paper we describe this type of search to identify all of the target genes in yeast that are regulated by the α2 and Mcm1 proteins that form a complex to bind DNA and repress expression of cell-type-specific genes. These experiments served to identify a new cell-type-specific gene in yeast. In addition, our results suggest that there are no extraneous α2–Mcm1 target sites in the genome and therefore all functional sites are regulating cell-type-specific functions.
RESULTS
Identification of Potential α2–Mcm1 Binding Sites
There are three different cell types in the yeast S. cerevisiae—haploid a, α, and diploid a/α cells—which differ in mating specificity and ability to sporulate. The differences in cell type are attributable to the expression of different cell-type-specific genes under the control of theMAT locus (Herskowitz 1988; Johnson 1992). TheMATα locus encodes α2, a homeodomain protein, which is required for transcriptional repression of a-cell-type-specific genes in α cells (Strathern et al. 1981; Tatchell et al. 1981;Herskowitz 1988; Johnson 1992). The α2 protein interacts with Mcm1, a MADS-box protein that is similar to the mammalian serum response factor, SRF (Keleher et al. 1988; Passmore et al. 1988; Treisman 1995;Tan and Richmond 1998). The α2 and Mcm1 proteins bind to a conserved, partially symmetric, 31-bp sequence located upstream of the transcription start site of a-specific genes (Johnson and Herskowitz 1985; Miller et al. 1985). The two proteins bind to the site as a heterotetramer, in which the Mcm1 dimer occupies the center region of the site and is flanked by α2 monomers. Although both proteins bind to this site on their own, they bind cooperatively with more than 100-fold higher affinity (Keleher et al. 1988). The Mcm1 protein also functions to set the spacing and orientation for α2 binding, which contributes to the specificity of the complex (Smith and Johnson 1992).
Previous studies have determined, in detail, the sequence requirements for the α2 protein to bind DNA with higher affinity and cooperatively in complex with Mcm1. Although each position in the 31-bp α2–Mcm1 recognition site has a base-specific preference, highly conserved TGTA sequences in each half site are essential for α2 binding and repression (Smith and Johnson 1994; Zhong and Vershon 1997). In the crystal structures of α2 binding alone and in complex with Mcm1, residues in the α2 homeodomain make specific contacts with these bases (Wolberger et al. 1991; Tan and Richmond 1998). In the center of the α2–Mcm1 site, the 5′-CCNNNNNNGG sequence has been showed to be required for Mcm1 binding (Treisman et al. 1989;Acton et al. 1997; West et al. 1997). The distance between the α2 sites and the Mcm1 site is variable among the natural half sites, with a spacing of 4 or 5 bp. Sites with 4 or 5 bp spacing repress transcription equally well, but sites with 3 or 6 bp spacing fail to repress transcription and are not bound cooperatively by α2 and Mcm1 in vitro (Mead et al. 1996). Although both α2 and Mcm1 make base-specific contacts to these bases, mutagenesis results show that there is relaxed sequence specificity, with a preference for A or T bases at these positions (Acton et al. 1997; Zhong and Vershon 1997). Taking the data from all of these experiments together, a degenerate consensus sequence of TGTAN(A/T)3–4CCN6GG(A/T)3–4NTACA can be deduced for the α2–Mcm1 binding site.
The degenerate consensus sequence was used to search for all the potential α2–Mcm1 recognition sites in the entire yeast genome using the PatMatch program provided by the SaccharomycesGenome Database. Allowing for a maximum of two mismatches in conserved bases, 59 sequences were obtained in the search (Table1). All of the sites that perfectly match the search sequence are located in the promoter regions of knowna-specific genes. Four sequences were identified which contain one mismatch with the search sequence. Two of these sites fall within the promoter regions between divergently transcribed open reading frames (ORFs) and therefore could potentially regulate both of these genes. Of the remaining 49 sites containing two mismatches with the degenerate consensus sequence, the majority of these are within ORFs. All of the previously identified α2–Mcm1 sites are within 250bp upstream of the translation start codon. It hasbeen shown that the sites placed further upstream of the gene or downstream of the start of transcriptionare significantly weaker in their ability to repress transcription (Johnson and Herskowitz 1985; A.K.Vershon, unpubl.). Therefore, because many of the sites in this group are within the genes and contain multiple mismatches at positions important for binding they are unlikely to be functional α2–Mcm1 repressor sites.
Potential α2/Mcm1–Binding sites in the Yeast Genome
Expression of Potential α-Specific Genes
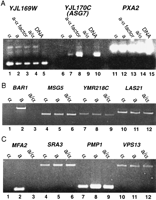
To determine if any of the α2–Mcm1 sites identified in the search regulate transcription of nearby genes, the expression of these genes was compared in different cell types by RT–PCR and Northern assays. Genes that are regulated by the α2–Mcm1 complex are repressed in α and diploid a/α cells and expressed ina cells.An example of the assay is shown in Figure1 and a summary of the results is shown in Table2. In addition to the known a-specific genes,STE2, STE6, MFA1, MFA2, BAR1, and AGA2, the only ORF that showed a similar pattern of expression in a versus α cells wasYJL170C. To confirm this result, the YJL170C coding region was used as a probe in a Northern blot of RNA prepared froma, α, and diploid a/α cells (Fig.2). Our results show that YJL170C is expressed in a but not in α or diploid a/α cells and is therefore an a-cell-type-specific gene. This is the seventh a-specific gene that has been identified and we have named this ORF ASG7.
(A) Expression of potential a-specific genes in haploid and diploid cells. The expression of YJL169W (lanes1–5), YJL170C (lanes6–10), and PXA2 (lanes11–15) were assayed using RT–PCR. RNA prepared from α (lanes 1,6,11), a (lanes2,7,12), a induced with α factor (lanes 3,8,13), and a/α diploid (lanes 4,9,14) strains was used as the template with primers specific for each ORF. Genomic DNA was used as the template to serve as controls for the PCR reactions and the size of the expected fragments (lanes 5,10,15). (B) Expression of BAR1 (lanes1–3), MSG5 (lanes 4–6),YMR218C (lanes 7–9), and LAS21(lanes 10–12) in α (lanes1,4,7,10), a (lanes 2,5,8,11), and a/α diploid (lanes 3,6,9,12) strains by RT–PCR. (C) expression of genes by RT–PCR that show cell-type-specific expression by microarray assay. MFA1 (lanes1–3), SRA3 (lanes 4–6),PMP1 (lanes 7–9), and VPS13 (lanes10–12) in α (lanes 1,4,7,10), a (lanes2,5,8,11), and a/α diploid (lanes 3,6,9,12).
Analysis of the Potential Sites for α2–Mcm1-Mediated Repression
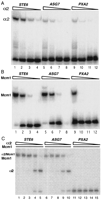
Expression of ASG7 (YJL170C) in haploid and diploid cells. Total RNA was prepared from MATα strain 246.1.1 (lane1), MAT a strain EG123 (lane 2), and the diploid MAT a/MATα strain 246.1.1 X EG123 (lane 3). The blot was hybridized with a radio-labeled DNA fragment specific for the coding region of the YJL170C. An ethidium bromide-stained gel before transfer is shown as a control for RNA loading.
The start of the coding region of the ASG7 gene is 397 bp away from the start of the YJL169W ORF. Because these genes are transcribed in the opposite direction, it is possible that they share promoter elements. In fact, the α2–Mcm1 binding site is closer to the start site of the YJL169W ORF than to ASG7. We did not see any evidence for a-specific expression of this gene by RT–PCR (Fig. 1) or Northern blot (data not shown). This result implies that there may be some asymmetry to the mechanism of regulation by this site.
Repression Mediated by α2–Mcm1 Target Sites of a Heterologous Promoter
Many of the sites that we identified do not appear to actively regulate gene expression in their endogenous contexts. It is still possible, however, that they are functional binding sites for the α2–Mcm1 complex and that they may serve a role in sequestering the α2 and Mcm1 proteins in the cell. To analyze whether the identified sequences are functional target sites for the α2–Mcm1 complex, we measured the level of α2–Mcm1-dependent repression mediated by the sites in the context of a heterologous promoter. Oligonucleotides containing the sites were cloned between the UAS and TATA elements in the promoter of a CYC1–lacZ reporter. The level of transcriptional repression mediated by each site was determined by comparing the levels of lacZ expression from constructs containing the potential α2–Mcm1 site to the parent vector without a site in a MATα strain (Table 2). All of the sites in the zero and one-mismatch groups, as well as sites that are most likely to be functional sites in the two-mismatch group, were assayed in the heterologous reporter vector. All of the previously identified sites found in the promoters of the a-cell-type-specificMFA1, MFA2, STE6, STE2, andBAR1 genes, as well as sites involved in determining the direction of mating type switching, DPS1 and DPS2, function as repressor sites in the heterologous promoter (Szeto et al. 1997; Zhong and Vershon 1997). The site in the promoter of thea-specific AGA2 gene, which was identified by our search and was previously uncharacterized for binding or repression, also functions as a strong repressor site in the heterologous promoter. The site upstream of ASG7 also mediates α2–Mcm1 repression of the test promoter, although the level of repression, 13-fold, is weaker than many of the sites found in othera-specific genes. All other sites tested do not show significant (>3-fold) levels of α2–Mcm1-mediated repression and are therefore not functional sites in vivo.
Mcm1-mediated Activation of the Potential Regulatory Sites
In the absence of α2, the Mcm1 protein binds to α2–Mcm1 sites and functions as a transcriptional activator (Bender and Sprague 1987; Jarvis et al. 1988; Ammerer 1990). To test whether any of the sites identified in the search function as Mcm1-dependent activator sites, the CYC1 UAS elements were removed from the heterologous reporters described above and lacZ expression was assayed in a MAT a strain (Table 2). All of the sites which function as repressor sites in the α strain also function as activator sites in the MAT a strain. With the exception of the site in the MSG5 promoter, most of the sites that do not repress transcription do not function as activator sites. TheMSG5 site, however, exhibits strong Mcm1-mediated activation in both a and α cells, suggesting that this site is a functional Mcm1 site.
In vitro Analysis of the AGA2 and ASG7α2–Mcm1 Binding Sites
The results described above show that our search has identified two previously uncharacterized, functional α2–Mcm1 target sites in theAGA2 and ASG7 promoters. To verify that the transcriptional repression and activation mediated by these sites is attributable to binding by the α2–Mcm1 complex, the sites were assayed for binding by α2 and Mcm1 alone, and in combination, by EMSAs (Fig. 3). The ASG7 site is bound by the α2 protein although the binding affinity is significantly weaker than that of the STE6 site. The Mcm1 binding affinity and the cooperative binding by α2 and Mcm1 to the ASG7site, however, are comparable with the levels observed to theSTE6 site. Under the same conditions, there is no detectable binding by the α2–Mcm1 complex to the site found in thePXA2 promoter. As expected from the in vivo repression data, the α2 and Mcm1 proteins have similar binding affinity for the site in the AGA2 promoter as to the STE6 site (data not shown). The in vitro results are therefore consistent with the in vivo results and show that the AGA2 and ASG7 sites function as α2–Mcm1 binding and repressor sites.
The DNA-binding activity for the ASG7 site in EMSAs. (A) Purified α2 protein was used in an EMSA with labeled fragments containing the indicated α2-Mcml-binding sites. The protein concentration was 4.0 × 10−7 m(lanes 1,5,9), 8.0 × 10−8 m (lanes2,6,10), 1.6 × 10−8 m (lanes3,7,11), 3.2 × 10−9 m (lanes 4,8,12). (B) Purified Mcm1 protein was used in an EMSA with labeled fragments at a concentration of 1.7 × 10−8 m (lanes1,5,9), 1.7 × 10−9 m (lanes2,6,10), 1.7 × 10−10 m (lanes3,7,11), and 1.7 × 10−11 m (lanes4,8,12). (C) Purified α2 protein at a concentration of 8.0 × 10−8 m (lanes 1,6,11), 1.6 × 10−8 m (lanes 2,7,12), 3.2 × 10−9 m (lanes 3,8,13), 6.4 × 10−10 m (lanes 4,9,14), 1.3 × 10−10 m (lanes 5,10,15) was mixed with Mcm11–96 fragment at a concentration of 1.7 × 10−9 m in the EMSAs. The α2–Mcm1 complex includes two molecules of α2 and two molecules of Mcm1 and the Mcm1 complex includes two molecules of Mcm1. the EMSAs shown are phosphorimages of the gels.
DISCUSSION
In this study, we have performed a search of the yeast genome for potential recognition sequences of the α2–Mcm1 complex. Assuming that the entire yeast genome has an A/T content of 61% (Dujon 1996) and allowing for two mismatches, we expected to find three to four matches in the genome to degenerate TGTAN(A/T)3–4CCN6GG(A/T)3–4NTACA sequence. The fact that we found 59 matches suggests that the presence of these sequences is not random. It is possible that a part of this sequence is used for recognition by other DNA-binding proteins or complexes. For example, some of the sites could have been identified because they contain an Mcm1-binding site. Mcm1 binds with other cofactors such as the α1, Ste12, Arg80, and Sff proteins to regulate other sets of genes (Lydallet al. 1991; Errede 1993; Hagen et al. 1993; Messenguy and Dubois 1993; Oehlen et al. 1996). One of the sites we identified is in the promoter of MSG5, a gene that is highly expressed in response to pheromone (Doiet al. 1994). We have shown that this site is not a functional α2–Mcm1 repressor site, but does serve as an Mcm1-dependent activator site and is likely required for the Ste-12–Mcm1 dependent pheromone response. Another explanation for the number of sites we found is that a large number of the sites are within the coding regions of genes or uncharacterized ORFs. Ten of the sites fall within the coding region of the PAU family of proteins, which have an unknown function and contain a low content of serine and threonine residues (Viswanathan et al. 1994). All of the potential α2–Mcm1 sites identified in this class of proteins fall in a highly conserved amino acid sequence. These sites were therefore identified in our search because they code for a conserved amino acid sequence and not because they are functional α2–Mcm1 binding sites.
Although the search identified a large number of potential sites in the genome, we have found that relatively few of these sites are functional α2–Mcm1 repressor sites. There appears to be a large decrease in the binding affinity and activity between the functional and nonfunctional sites and there do not appear to be any sites with intermediate affinity or activity. Most of the sites identified in the search contain two significant mismatches with the search sequence. With the exception of the DPS2 site, all show substantial decreases in binding affinity in vitro and repression of a heterologous promoter in vivo when compared with the functional sites. In addition, none of the genes near these sites show significant cell-type-specific expression in DNA microarray experiments (Roth et al. 1998). These results further support our findings that these are not functional regulatory sites. It is unlikely that these sites are bound by the protein in vivo. As a result, there do not appear to be any active α2–Mcm1 sites in the genome without a specific regulatory target and the presence of an active α2–Mcm1 site indicates a functional target gene.
Our search identified two previously uncharacterized α2–Mcm1 target sites. One is upstream of AGA2, a cell-type-specific subunit of the a-agglutinin cell adhesion complex (Cappellaro et al. 1994). Northern results demonstrated that the AGA2 gene is expressed specifically in a cells (de Nobel et al. 1995). Although we expected this gene to be regulated by α2–Mcm1, the promoter of this gene had not been analyzed previously. From the search for potential α2–Mcm1 sites, we obtained a sequence in the promoter region of AGA2 that matches the consensus α2–Mcm1 site. Furthermore, we have shown that it is a functional α2–Mcm1 site in vivo and in vitro. Repression mediated by this site is as strong as α2–Mcm1 sites in other acell-type-specific genes (Table 2) and the α2 and Mcm1 proteins bind to the AGA2 site with the same affinity as to theSTE6 operator (data not shown). Therefore, as expected, expression of the a cell-type-specific AGA2 gene is directly regulated by the α2–Mcm1 repressor complex.
The other novel α2–Mcm1-binding site identified in our search is located 205 bp upstream of the translation start site ofYJL170C. This gene is only expressed in a cells and is strongly induced by exposure of the cells to α-factor (Fig. 1).YJL170C is therefore a novel a cell-type-specific gene that we have named ASG7. A chromosomal null mutation ofASG7 does not have an effect on the growth rate or mating efficiency (H. Zhong, unpubl.). Preliminary results have shown, however, that ASG7 is required for inhibition of thea-factor mating type receptor, Ste-3 (J. Kim and J. Hirsch, pers. comm.).
Knowledge of the yeast genome sequence makes it possible to examine the expression profile of every gene in the genome under different conditions using DNA microarray techniques (DeRisi et al. 1996; Chu et al. 1998; Roth et al. 1998). This type of analysis has been performed to examine the differential expression of genes in a and α cells (Roth et al. 1998). In agreement with our results and previous findings by others, MFA1, MFA2, AGA2,STE2, and BAR1 show significant differences in expression in a versus α cells. Several other genes,PMP1, SRA3, and VPS13, which we did not identify in our search, also show higher expression in a versus α cells in the microarray assay. We did not find any potential α2–Mcm1 binding sites in the promoter of these genes, however, and did not observe differential cell-type-specific expression of these genes in our assay (Fig. 1C). Two of the genes identified in our search, STE6 and ASG7, did not have large enough differences in the microarray assay to be identified as cell-type-specific genes. Furthermore, the DPS1 andDPS2 sites, which have α2–Mcm1-dependent roles in the direction of mating type switching, would not be detected in the microarray assay. These sites regulate the cell-type-specific expression of several small transcripts in the left arm of chromosome III that are unlikely to code for any functional protein (Szeto et al. 1997). These transcripts would therefore not likely be detected in microarray assays that monitor expression of well-defined ORFs. Taken together, these results show that each technique identified potential target genes that were not detected by the other assay and that together the two techniques complement. In summary, the search we have performed has identified two previously unidentified sites as well as all of the known α2–Mcm1 regulatory sites. This work gives an example that, if the sequence requirement of a DNA-binding protein is relatively well defined, it is possible to search a genome database for its target sites and therefore find novel target genes that are regulated by this protein.
METHODS
Northern and RT–PCR Assays
The total RNA from strains 246.1.1 (Mata), EG123 (Matα), and 246.1.1 X EG123 (Mata/α) (Silicano and Tatchell 1984) was prepared as described (Ausubel et al. 1997). RNA from a cells treated with α-factor was purified in the same manner except that cells were harvested 2 hr after adding 2 mm of α-factor at OD600 of 0.3. RT–PCR assays were performed using the SuperScript Preamplification System kit (GIBCO BRL) according to the manufacturer's instructions. Two micrograms of total RNAs isolated from a, α, or a/α yeast strains were used as templates and oligo(dT)12–18 was used as the primer to synthesize cDNAs. Pairs of oligonucleotides that specifically anneal to each of the potential genes were used as primers in the second step of PCR for amplification of target cDNAs. These primer pairs were designed to generate ∼500-bp fragments. The reactions were run on agarose gels and DNA fragments were visualized with ethidium bromide.
For Northern blot analysis, RNA samples were electrophoresed on a 1% formaldehyde–agarose gel and the RNAs were denatured, neutralized, and blotted to a nitrocellulose membrane. The level of mRNA expression was detected by hybridization with PCR-generated fragments of each gene, which were labeled by random priming with [α-32P]dCTP. Hybridization was performed at 42°C in buffer containing 50% formamide and washed in 2× SSC (Herrick et al. 1990). The membrane was exposed to a phosphor screen and the image was scanned on a Molecular Dynamics phosphorimager.
β-Galactosidase Assays
Transcription reporter plasmids that contain potential α2–Mcm1-binding sites were constructed by inserting double-stranded oligonucleotides containing the 31- or 32-bp sites with TCGA overhangs into the XhoI site between the UAS and TATA elements in the CYC1–lacZ promoter of pTBA23 (Mead et al. 1996). These constructs were used to measure α2–Mcm1-dependent repression of the CYC1 promoter. Plasmids used to measure Mcm1-dependent activation were constructed by digesting the above plasmids with BglII, followed by self-ligation (Acton et al. 1997). This step removed the CYC1 UAS sites from the promoter region, leaving the α2–Mcm1 site and CYC1 and TATA box. The reporter constructs were transformed into the MATα strain 246.1.1 to assay repression or the MAT a strain EG123 to assay activation by measuring the level of β-galactosidase activity as described previously (Silicano and Tatchell 1984; Keleher et al. 1988).
EMSAs
Full-length α2 and a fragment of Mcm1 (residues 1–98) were expressed in bacteria and purified to >90% homogeneity as described previously (Mead et al. 1996; Acton et al. 1997). The relative α2–Mcm1 DNA-binding affinity for the potential α2–Mcm1-binding sites were determined by EMSAs as described (Zhong and Vershon 1997).
Acknowledgments
We thank J. Gu for help in constructing some of the LacZreporter plasmids and J. Kim and J. Hirsch for communication of unpublished work on the function of ASG7. This work was supported by a grant from the National Institutes of Health to A.K.V. (GM49265).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵1 Corresponding author.
-
E-MAIL vershon{at}mbcl.rutgers.edu; FAX (732) 445-5735.
-
- Received March 24, 1999.
- Accepted September 3, 1999.
- Cold Spring Harbor Laboratory Press