Caenorhabditis elegans Has Scores of hedgehogRelated Genes: Sequence and Expression Analysis
Abstract
Previously, we have described novel families of genes, warthog(wrt) and groundhog (grd), inCaenorhabditis elegans. They are related to Hedgehog (Hh) through the carboxy-terminal autoprocessing domain (called Hog or Hint). A comprehensive survey revealed 10 genes with Hog/Hint modules in C. elegans. Five of these are associated with a Wart domain in wrt genes, and three with multiple copies of the Ground domain in grd genes. Both the Wart domain and the Ground domain occur also in genes encoding no Hog domain. Further, we define a new group of genes related to the grd genes, calledground-like (grl). Overall, C. elegans has more than 50 genes belonging to these gene families. Phylogenetic and sequence analysis shows that the wrt, grd, and grlgenes are derived from each other. Further examination reveals a sequence motif with similarity to the core of the amino-terminal-signaling domain of Hh proteins. Our data suggest that the wrt, grd, grl, and hh genes are derived from a single ancestral gene. wrt, grd, and grl genes are also present in other nematodes, but so far not in any other phyla. Conversely, hh is not found presently in C. elegansnor other nematodes. Thus, the nematode genes could be the homologs of Hh molecules in other phyla. The membrane molecule Patched has been shown previously to be a receptor of Hh. Many Patched-related proteins are present in C. elegans, which may be targets of thehh-related genes. No Hedgehog-interacting protein (Hip) was found. We analyzed the expression patterns of eight wrt and eight grd genes. The results show that some closely related genes are expressed in the same tissues, but, overall, the expression patterns are diverse, comprising hypodermis, seam cells, the excretory cell, sheath and socket cells, and different types of neurons.
[Hog domain-containing genes are available as an online supplement table at www.genome.org. The sequence data described in this paper have been submitted to the GenBank data library under accession nos.AF139522, AF139520, AF139521]
hedgehog (hh) genes encode a family of secreted signaling molecules with functions in anteroposterior patterning as well as differentiation of neurons and many other cell types in flies and vertebrates (for review, see Hammerschmidt et al. 1997). One member of the hh gene family has been identified in Drosophila, whereas multiple genes are known from vertebrates that arose during chordate evolution (Zardoya et al. 1996). All of these genes encode a highly conserved amino-terminal signaling domain that is cleaved off and anchored to a cholesterol moiety by the protein's own carboxy-terminal protease domain (Porter et al. 1996b). The biological function of the cholesterol anchor is presently unknown (for review, see Goodrich and Scott 1998).
The Caenorhabditis elegans genome sequencing project had revealed several genes and ESTs that encode a Hh-like autocleavage domain at their carboxyl terminus (Bürglin 1996;Porter et al. 1996a). We called this domain the Hog domain, because of the sequence similarity to Hedgehog (Bürglin 1996); the part that shares functional and structural similarity with intein domains in prokaryotes is also referred to as the Hint domain (Hedgehog/intein; Hall et al. 1997). TheC. elegans genome sequence is now essentially completed (TheC. elegans Sequencing Consortium 1998), however, none of the Hog domain-encoding genes are obvious hh homologs. The region amino-terminal to the Hog domain of several of these genes encode two novel domains we termed Wart and Ground (Bürglin 1996). Genes that encode only the new domains but lack the Hog domain have also been found. These gene families have been named warthog (wrt) andgroundhog (grd), irrespective of their association with a Hog domain. The Wrt and Grd molecules have, in many cases, good signal sequences for protein export at their amino termini, therefore they are probably secreted. Porter et al. (1996a) have shown that the ZK1290 protein (Wrt-1) cleaves itself as predicted when expressed inDrosophila cell culture cells.
In this work we present a complete survey of all wrt, grd, and related genes found in the C. elegans genome. In addition, we have investigated the expression pattern of previously described genes (wrt-1 to wrt-8, grd-1 to grd-3, andgrd-5 to grd-9 (Bürglin 1996). We have also cloned some new cDNAs for wrt genes to confirm the ORF predictions and to demonstrate transcription. Further, our sequence and phylogenetic analyses indicate that the amino-terminal domains and the carboxy-terminal Hog domain coevolved during evolution so that all of the nematode genes derive monophyletically from the same common ancestor as the hh genes.
RESULTS
Database Searches
Our previous analysis (Bürglin 1996) was limited by the unfinished state of many sequences. The availability of the almost complete genomic sequence of C. elegans makes it now possible to produce a comprehensive overview of all of the genes containing a Hog domain and other motifs found in association with the Hog domain. All sequences presented here are predictions based on our ORF analysis, taking into account homology, cDNAs (ESTs), and signal sequences for protein export. Our reassessment showed that some of our ORF predictions, as well as many from the sequencing consortium, needed corrections.
During our searches, we also found homologs for the different gene families in other nematode species. Several genes were found in the genome of Caenorhabditis briggsae, ∼5% of which is sequenced (Genome Sequencing Center, St. Louis, pers. comm.). This nematode is closely related to C. elegans (Blaxter et al. 1998) and has served to identify coding and regulatory regions; generally, protein coding regions are highly conserved between these two species (for review, see Sluder et al. 1999). C. briggsaesequences thus serve to confirm C. elegans ORFs as real genes rather than pseudogenes. Several genes were also found in the EST projects of the parasitic nematodes Brugia malayi andOnchocerca volvulus (Blaxter et al. 1996, 1999; The Filarial Genome Project 1999), both members of the order Spirurida. These filarial parasites are very divergent from C. elegans (Blaxter et al. 1998) and are estimated to have diverged some 400 million years ago (D. Fitch and M. Blaxter, pers. comm.). Genes that can be identified as clear orthologs have thus been conserved over a long period of time, whereas genes that have no clear orthologs indicate more rapid divergence and diversification.
Hog/Hint Domains in the C. elegans Genome
BLAST searches, PSI–BLAST searches (see Methods), and subsequent comparisons revealed 10 Hog domains in the genome of C. elegans. This contrasts with a previous, more global survey that reported 11 Hint modules (Chervitz et al. 1998); two of these encode only a Wart domain, whereas one Hog domain was missed. Although our PSI–BLAST searches did detect many prokaryotic inteins, apparently no inteins are present in C. elegans. Except for one Hog domain (W06B11.4), all are associated with amino-terminal sequences that have signal sequences for protein export (Bürglin 1996, Fig. 1, 6C). Five of the Hog genes arewrt genes (wrt-1, wrt-4, wrt-6, wrt-7, andwrt-8) and three are grd genes (grd-1, grd-2, and grd-11). The gene M110 has a long ORF upstream of the Hog domain. Approximately the first 250 residues show a unique, diverse amino acid composition, whereas the central region consists of repeated amino acids. Searches with the amino-terminal region revealed no matches to any other protein, thus, M110is an orphan hog gene. The gene W06B11.4 encodes only a Hog domain; its first methionine starts within the Hog domain. However, the similarity can be extended on the genomic sequence up the cysteine residue at the cleavage site (Fig. 1A), but no splice site and no other methionine can extend the ORF further. Thus, W06B11.4 is probably a pseudogene. In conclusion, the 10 Hog domains are encoded by several gene families: 5 wrt, 3grd, 1 orphan, and 1 hog only. The online supplement table lists all of the Hog domain-containing genes as well as all of the associated and derived motifs.
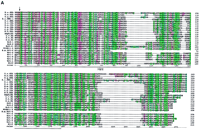
Sequence alignments of the Hog/Hint domains and associated motifs. The color coding of the residues is slightly modified from the default settings in ClustalX. In particular, small hydrophobic residues are green, large hydrophobic residues are dark green, and cysteine residues are yellow in all positions. In B–D the residues of the core motif (C-FDφV-C, see text) are shown above the alignments; the line under the first Cys in C indicates a variable position, small c in D indicates additional conserved residues. (A) Alignment of Hh Hog domains and the nematode Hog domains. (Arrow) The site of autocatalytic cleavage. A line marks the definition of the Hint domain. The whole alignment was used for the neighbor joining (NJ) analysis in Fig. 2A. Sequences used were taken from Bürglin (1996) with the addition of sea urchin (L.v.Hh,Lytechinus variegatus, GenBank accession no. AF059606), Amphioxus (AmphiHh, Y13858), newt (C.p.Hh, Cynops pyrrhogaster, D63339), and Drosophila hydeii (D.h.Hh, AAB34104). Other species codes are: (H.s.) human; (M.m) mouse; (G.g.) chicken; (D.r.) zebrafish; (X.l.) Xenopus laevis; (D.m.)Drosophila melanogaster. (B) Alignment of the Wart domains. The region use for the NJ analysis is marked by a line. (C) Alignment of the Ground domains. (rp) Repeat number. Beneath the secondary structure, prediction for the alignments is indicated. Squiggles mark loop regions, green bars α-helixes, and zigzag lines β-strands. Y102A5c.34 needs two frameshifts near the amino terminus—marked with a small x—to line up all the residues; Y69A2A.x has also some problems with extending the ORF over the conserved region, although this YAC sequence is still unfinished. (D) Alignment of the Ground-like domains. Secondary structure predictions as for C. The reading frame for T24A6.3 was changed by hand to get a good methionine with a signal sequence. T24A6.x seems to contain only the last two-thirds of the Ground-like domain. The sequence similarity of C.b.F32D1.4 extends over only one exon, with no other similarities in the flanking regions to be found.C.b.F32D1.4 might be a recent pseudogene, suggesting thatF32D1.4 may not be highly conserved in evolution.
Sequence comparison of the nematode Hog domains shows that they can be aligned with the carboxy-terminal domain of the Hh proteins throughout the entire length extending past the Hint domain (see Fig. 1A). The similarity in the region past the Hint domain is primarily confined to hydrophobic residues that occur in conserved intervals. In Hh proteins, this region has been identified as a sterol recognition region (SRR;Beachy et al. 1997; Hall et al. 1997); however, it is unknown whether the carboxyl termini of the C. elegans Hog domains also recognize sterols.
Redefinition of the Wart Domain
Five of the Hog domain genes encode an amino-terminal motif that we termed Wart domain (Bürglin 1996). Searches with this motif revealed five additional genes in C. elegans encoding only the Wart domain as a conserved motif. These 10 genes together have been named wrt genes. Nine of the wrt genes,wrt-1 to wrt-9, encode a Wart domain that is readily recognizable (Fig. 1B). wrt-10 is highly divergent and scored only very low in BLAST searches. It lies on cosmid ZK1290 right next towrt-1 on the opposite strand (Fig. 3, below). Despite the sequence divergence, Wrt-10 shares all the conserved cysteine residues as well as other residues, particularly in the carboxy-terminal region of the Wart domain. An ortholog for wrt-2 was found in C. briggsae. One wrt gene, B.m. wrt-6, apparently a homolog of wrt-6, was found among the B. malayi ESTs. Curiously, this EST encodes only the approximately first half of the Wart domain and then the EST continues immediately into the Hog domain (Fig. 1A,B,5C). This cDNA could thus either represent a divergent form of a wrt gene, or an alternative splicing product. Orthologs for the divergent wrt-10 gene were found both in ESTs fromB. malayi and O. volvulus. Database searches did not reveal any Wart domains in phyla outside of the nematoda.
Genomic structure and reporter constructs. ORFs are indicated above the chromosome, represented by the different cosmids. ORFs in black aregrd and wrt genes, gray ORFs are neighboring genes, small gray ORFs beneath grd or wrt genes are the computer predictions from ACeDB. Fragments used for reporter constructs are drawn above the respective ORFs: (gfp) Hatched boxes; (lacZ) dark gray boxes. wrt-7 and wrt-8 are separated from each other by two intervening ORFs, which are themselves a tandem duplication. Upstream of wrt-8, two otherH02K04-1A related ORFs are also present, indicating that a duplication of this whole gene cluster occurred.
Our reassessment of the wrt genes extends the Wart domain at the carboxyl terminus, revealing one additional conserved cysteine residue (Fig. 1B). The consensus Wart domain thus has eight conserved cysteine residues.
Analysis of Ground Domain Genes Reveals Flexible Patterns of Cysteine Conservation
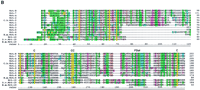
A second family of Hog-encoding genes are the grd genes. Three of these genes, grd-1, grd-2, and grd-11 encode a Hog domain. Sequence comparison shows that these genes are closely related to each other over their full length, each encoding four Ground domains amino-terminal to the Hog domain (see online supplement). In all three genes, the Hog domain is separated from the fourth Ground domain by variable lengths of homopolymeric amino acid tracts. Interestingly, two positions in the fourth Ground domain of Grd-11, in which conserved cysteine residues are found, have been replaced by other residues.
Searches with the Ground domain revealed 14 additional genes in C. elegans, each having only one Ground domain (Fig. 1C). These genes are usually rather small, although in some cases repetitive regions separate the signal sequence for protein export from the Ground domain (Bürglin 1996). Two of the genes may be pseudogenes,Y102A5c.34 and Y69A2A.x. A Ground domain-encoding gene has been found in the B. malayi EST project (Fig. 1C), similar to grd-5 and grd-10. No other Ground domains have been found in the databases.
Several of the C. elegans grd genes are clustered on the chromosomes. One gene cluster, grd-3, consists of grd-13, grd-3, and grd-10; the second, grd-4, consists ofgrd-14 and grd-4 (Fig. 3, below). Both Grd-13 and Grd-14 display two differences compared to other Grd proteins; the second cysteine residue of a conserved doublet has been changed to another residue (Fig. 1C). Concomitantly, whereas all of the other Ground domains encode a cysteine residue 9–19 residues upstream of the cysteine doublet, Grd-13 and Grd-14 have no such residue. We conclude that a typical Ground domain consists of four cysteine residues, two of which are in an adjacent doublet, and that Grd-13 and Grd-14 lost one pair. Given that we see several instances of pairwise loss of cysteine residues, it is tempting to infer that disulfide bonds are formed by such pairs. A possible arrangement of disulfide bonds in shown in Figure 5A, below.
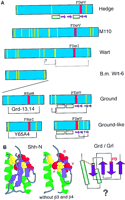
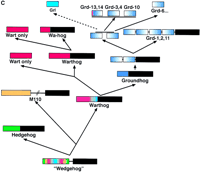
Hypothesis for the structure and evolution of the hh, wrt, grd, and grl genes. The green bars denote α-helixes, and the purple arrows β-strands in A and B. (A) Schematic representation of the different conserved domains and the amino-terminal region of M110. The yellow lines indicate conserved cysteine residues. The red box indicates the conserved FDφV motifs found in variations in all sequences. The brackets indicate putative cysteine disulfide bonds. Underneath the several domains are schematic representations of the secondary structure (Hedge) or secondary structure predictions (Grd, Grl). (B) The central part of the Shh-H structure viewed with RasMol; (right) β-strand 3 and 4 were removed. The FDφV motif is represented in red, the cysteine residues in yellow. The scheme at right is a topologically allowed scheme of how the Ground and Ground-like domain could be arranged. (C) Evolution of the hog gene families from a common ancestralwedgehog gene. Genes are represented schematically; the regions between the domains not drawn to scale. The black box denotes the Hog region; the other colors the different amino-terminal domains, i.e., Hedge, Wart, Ground, and Ground-like.
A New Group of Genes Related to grd Genes:ground-like Genes
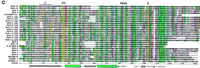
During the BLAST and PSI–BLAST searches, additional ORFs were detected that display low sequence similarity to the Ground domain. We refer to this new group of genes as ground-like(grl). Twenty-eight grl putative ORFs were discovered. They all share features in common with Ground domain genes as follows: they have signal sequences for protein export, often stretches of repetitive amino acids separate the signal sequence from the Ground domain, and the genes are in general rather small like thegrd genes. Most terminate right after the Ground-like domain. Despite the low primary sequence similarity, significant scores between the Ground and the Ground-like domains are achieved: PSI–BLAST searches for Ground domains in GenBank revealed all Ground domain genes, as well as Grl F32D1.4 with a probability of 2e−14. BLAST searches at the Sanger Center with Grl F42C5.7 revealed many Grl ORFs, but also Grd-10 and Grd-5 with probabilities of 1.2e−6 and 6.1e−6, respectively.
The grl genes are highly divergent (Fig. 1D). On cosmid T24A6, five regions of similarity could be detected; however, two may be pseudogenes. Five grl genes were found in the C. briggsae genomic sequences, and all could be assigned as orthologs of C. elegans genes (Fig. 1D), although one (C.b.F32D1.4) might be a pseudogene. Two grl genes were found in the EST project of B. malayi.
The grl genes distinguish themselves from the grdgenes by different patterns of conserved cysteine residues. Instead of the cysteine doublet, only one cysteine residue is conserved, and no cysteine residue is present upstream, which is reminiscent of Grd-13 and Grd-14. In contrast, the grl genes encode two new conserved cysteine residues at other positions, one in the middle and one toward the carboxyl terminus of the Ground-like domain (Figs. 1D and 5A, below). In the case of Grl ORF Y65B4.x, this latter pair of cysteine residues is not present. Gain and loss of pairwise cysteine residues again suggests that a disulfide bridge may be formed (Figs. 1D and 5A, below).
Phylogenetic Analysis Indicates Proliferation and Diversification of wrt, grd, and grl Genes
Wart Domain
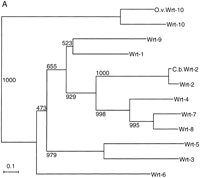
To determine the evolutionary history of these gene families, we have performed phylogenetic analysis of the various domains with neighbor-joining (see Methods). The Wart domain tree (Fig.2A) shows clear clustering of wrt-3 andwrt-5 as well as of wrt-7, wrt-8, and wrt-4. Further, wrt-2 forms a clade with wrt-4, wrt-7, andwrt-8, and the C. briggsae wrt gene is the ortholog of wrt-2. The close similarity of wrt-7 andwrt-8 is confirmed by the chromosomal clustering of these two genes. The B.m. wrt-6 gene could not be classified on the basis of its partial Wart domain, which mostly did not cluster with any particular wrt gene. The most divergent wrt gene,wrt-10, forms a distinct clade with homologs from B. malayi and O. volvulus, indicating long-time conservation of this gene (Fig. 2A).
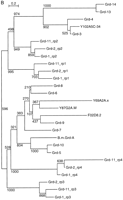
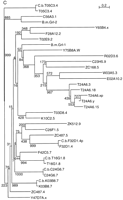
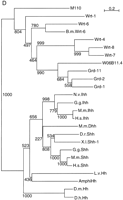
Evolutionary dendrograms of the various domains. Trees were generated by the neighbor joining method, numbers indicate the bootstrap values. A small p behind the name marks a possible pseudogene. (A) Dendrogram of Wart domains. (B) Dendrogram of Ground domains. (C) Dendrogram of Ground-like domains. (D) Dendrogram of the Hog domains.
The presence of two distinct types of wrt genes in B. malayi suggests that several wrt genes were already present before the separation of Spirurida from Rhabditida. Given thatwrt-10 and wrt-1 are clustered, and thatwrt-10 has been highly conserved in evolution suggests a very early duplication from wrt-1. It also implies that thewrt-1 gene in this cluster may have served as the original source for the radiation of the remaining wrt genes. Interestingly, the tree also indicates that the Hog domain has been lost several times independently during the evolution of thewrt genes as genes encoding no Hog domain are found in several places within the tree (Fig. 2A).
Ground Domain
The Ground domain genes can be grouped into distinct subfamilies (Fig. 2B). The Ground domains of grd-1, grd-2, andgrd-11 build distinct subgroups, suggesting that these genes duplicated only recently. Those genes encoding only a single Ground domain fall into two separate clades, one well-defined clade withgrd-3, grd-4, grd-13, grd-14, and Y102A5c.34 and another with the remaining grd genes. The grd-3 andgrd-4 gene clusters (Fig. 3) are clearly derived from each other. grd-13 and grd-14 form a significant clade with grd-4 and grd-3, which shows that grd-13 and grd-14 are duplications deriving fromgrd-3 and grd-4, respectively, as their position on the chromosome suggests (Figs. 2B and 3). grd-5 andgrd-10 are also closely related to each other (Fig. 2B). Given that grd-10 is part of the grd-3 cluster,grd-5 was probably originally part of the grd-4cluster. The B. malayi grd gene B.m. grd-A forms a clade with both grd-5 and grd-10 indicating it is a homolog of a common ancestor. It suggests that the duplication ofgrd-5 and grd-10 and the grd-3 andgrd-4 clusters happened after the divergence of Spirurida and Rhabditida.
What is the relationship between thegrd-1/grd-2/grd-11 and the Ground only encoding genes? The problem depends on the placement of the root in the tree in Figure 2B. However, we can also take flanking sequences into account. The complete alignment of Grd-1, Grd-2, and Grd-11 shows that the first and the second Ground domains are duplicates of each other as the sequence similarity can be extended well past the Ground domain (online supplement). This extended sequence similarity is also found in Grd-3, Grd-4, Grd-13, and Grd-14. We conclude from this observation that the latter genes are more closely related to repeat 1 or 2 of an ancestral Grd protein (Fig. 5C). Conversely, the phylogenetic analysis suggests that Grd-5 to Grd-10 and related proteins are derived from repeat 4 of an ancestral Grd protein. Because grd-3 and grd-10are in a cluster, we have the same arrangement of the Ground domains as in grd-1/grd-2/grd-11. On the basis of additional data (see below), we infer that an ancestral grd gene with two to four copies of the Ground domain duplicated and the duplicate converted into the ancestor of the grd-3/grd-4 gene cluster and lost the Hog domain (Fig. 5C). The ancestral grd-5/grd-10 gene then gave rise to the additional grd genes such as grd-6 togrd-9, which suggests a rapid expansion. This is supported by the fact that 8 of 17 grd genes are found on chromosome V, which harbors many expanded gene families, for example, nuclear receptors and G-coupled receptors (Bargmann 1998; Sluder et al. 1999).
Ground-Like Domain
An analysis of the grl genes did not reveal any special subgroupings of the diverse genes (Fig. 2C); only a few genes fall into defined subfamilies, such as those on cosmid T24A6. Otherwise, the branch points are deep and poorly resolved. Even the genesZC487.4 and ZC487.5, which lie next to each other, do not form a clade. The five C. briggsae grl genes can all be assigned as obvious orthologs of C. elegans genes (Fig. 2C), suggesting at least that these genes have been subject to evolutionary constraint. Of the two B. malayi genes, one (B.m. grl-2) clusters with C56A3.1 and is very likely the ortholog, but the other (B.m. grl-1) cannot be assigned as a homolog of any particular C. elegans grl gene (Fig. 2C). These data suggest a rapid proliferation and diversification of thegrl genes, which is also supported by the observation that 19 of 28 genes are found on chromosome V. Conversely, grl genes were already present before the divergence of Spirurida and Rhabditida.
Hog Domain
Phylogenetic analysis of the Hog domain shows that the hhgenes form a distinct clade (Fig. 2D). A comparative tree generated with Pileup gives a virtually identical branching pattern (data not shown). Furthermore, on the basis of the primary sequences of the Hog domain, especially the carboxy-terminal region (Fig. 1A), the nematode domains share more characteristics with each other than with the domains of Hh. Thus, we think the placement of the root of the tree is a good reflection of the true situation.
grd-1, grd-2 and grd-11 are closely related to each other (Figs. 1C and 2D). W06B11.4 seems to be derived from grd genes, but presumably lost the Ground domain-coding regions. Thus, W06B11.4 cannot be a remnant of a now lost hh gene. wrt-4, wrt-7, andwrt-8 form a clade, as also predicted by the Wart domains (Fig. 2A,D). The partial Hog sequence of B.m. wrt-6 clusters with wrt-6, which lead us to classify it as a homolog ofwrt-6. Of the wrt genes, wrt-1 seems to be the most ancestral one, which supports the idea that wrt-1 andwrt-10 form an ancient cluster. Importantly, the tree indicates that the Hog domain of the grd genes is located within that of wrt genes. The orphan hog geneM110 seems to take a most divergent position within theC. elegans genes, consistent with the fact that the amino terminus shares no obvious sequence similarity with other genes. Overall, none of the C. elegans Hog domain genes are in any way more closely related to the hh genes.
The Amino-Terminal Domains Share a Similar Core Sequence
The Hog domain is related to intein molecules, a group of self-splicing mobile elements (Cooper and Stevens 1995; Perler et al. 1997). This raises the issue of whether the Hog domain has been acquired multiple times independently by different genes, e.g., theM110, wrt, and grd genes. However, the phylogenetic analysis indicates that the Hog domain of the grd genes is derived from that of the wrt genes (Fig. 2D). Therefore, we examined the Wart, Ground, and Ground-like domains for similar features.
Conserved features of the second part of the Wart domain are a cysteine residue, a cysteine doublet, a motif consisting of the four residues (F or Y), (D), (φ = hydrophobic), (I or V), and another cysteine residue (Figs. 1B and 5A). Likewise, the Ground domain contains a conserved cysteine doublet and a single cysteine residue that flank a central motif (F, Y, or A), (E or D), (φ, S, or T), (V or I) in a spacing similar to that in the Wart domain (Figs. 1C and 5A). The Ground-like domain also shares similar features with the Ground and Wart domain, although the pattern of conserved cysteine residues has undergone changes, as shown above (Figs. 1D and 5A), and the central core motif is less conserved, perhaps not surprising given the proliferation of grl genes. On the basis of these observations, we deduce that the Ground domain is derived from the second part of the Wart domain, and that a wrt gene consisting of a Wart and a Hog domain gave rise to an ancestral grd gene (Fig. 5C).
Extending the analysis to M110, we also find a cysteine doublet upstream of an F, D, G, V sequence (Fig. 5A), which is followed by a cysteine residue, although we cannot judge the conservation of this pattern, because there is only one sequence. Extra cysteine residues are present within this region of M110, however, this is not unusual as extra cysteines are also found in the Ground-like domain. Examination of the amino-terminal sequences of Hh proteins—the Hedge domain—(Hall et al. 1995) also reveals an absolutely conserved F, D, φ, V motif. This motif is flanked upstream by a conserved cysteine and downstream by another cysteine, although the latter is not conserved in the Desert Hedgehog family (Zardoya et al. 1996). Examination of the structure of the amino-terminal domain of Sonic Hedgehog (Shh-N; Hall et al. 1995) reveals that the motif is located in the central core of the protein. The presence of a conserved motif that we will refer to as C-FDφV-C suggests that the amino-terminal regions of the hh, M110, wrt, grd, and grl genes diverged from a single common ancestor.
Transcription of wrt, grd, and grl Genes and Reporter Construction
cDNAs from EST projects have been identified for many of thewrt, grd, and grl genes. To obtain additional cDNAs, we performed PCR using either a cDNA library or first strand cDNA, and cloned and sequenced cDNAs for wrt-3, wrt-5, andwrt-6. For wrt-7, we were not able to obtain a cDNA. Thus, 8 of 10 wrt genes are confirmed to be expressed. In particular, wrt-10 seems to be highly expressed, because many ESTs were recovered not only from C. elegans, but also from the parasitic species O. volvulus and B. malayi.
Of the grd genes, at least five are also expressed on the basis of the ESTs. For 9 of the 28 grl genes, ESTs have been found, indicating that many of them are also transcribed. Several of the grl genes have C. briggsae orthologs, which suggests that they are actively transcribed genes under evolutionary constraint. The orphan hog gene M110 is also transcribed.
Overall, there is convincing evidence that at least 26 of the 57 genes are expressed. Conversely, seven genes are possibly pseudogenes, because of sequence abnormalities or lack of expression. Thus, we are confident that there are between 30 and 50 bona fide hhrelated genes in C. elegans.
Using start codon::gfp fusions we attempted to determine the gene expression patterns of wrt-1 to wrt-8 (wrt-nM::gfp), grd-1 to grd-3, and grd-5 to grd-9 (grd-nM::gfp). As several grd genes had ambiguous start positions on the basis of the initial sequence analysis (Bürglin 1996) and cDNAs are not available, we also fused gfp–lacZreporters (grd-nC::gfp–lacZ) into the conserved region of the Ground domain (Fig. 3).
Expression Pattern of wrt Genes
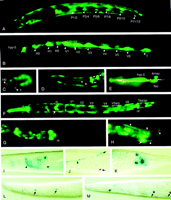
All wrt-nM::gfp fusions are expressed to various extents in hypodermal syncytia, except wrt-7M::gfp, which is not expressed at all. Three constructs, wrt-1M::gfp, wrt-4M::gfp, and wrt-8M::gfp, are expressed strongly and exclusively there. The other wrt-nM::gfp constructs stain the hypodermis weakly and are expressed most strongly in other tissues (see below). Earliest GFP expression is detectable in the seam cells of 1.5- to 1.8-fold embryos (wrt-2, wrt-5), other expressions have their onset at the 3-fold embryonic stage.
wrt-1M::gfp, wrt-4M::gfp, and wrt-8M::gfp are all expressed in the hypodermal syncytia hyp6 to hyp10 from threefold stage to adult embryos (Fig. 4A; data not shown).wrt-8M::gfp expression is stronger in anterior and posterior syncytia (data not shown). Additionally, wrt-8M::gfp is expressed in the 12 subventral P cells from the 3-fold embryonic stage to early L2 larvae (Fig. 4A). We can follow GFP expression during ventral migration of P cells and in some of their hypodermis and neuroblast descendants until the early L2 stage. Expression continues after fusion with the hypodermal syncytium but is not found in P3.p to P8.p that form the vulval equivalence group.
Expression patterns of wrt and grd reporter constructs. Worms are oriented with anterior to the left and ventral downward unless indicated otherwise. (A) Fluorescent micrograph of L1 larva: wrt-4M::gfp is expressed in hypodermal syncytia and the subventral P blastomeres. (B,C) wrt-2M::gfp expression in all seam cells of an L1 larva (B) and an 1.8-fold stage embryo (C). (D) Head of L3 larva. wrt-3::gfp is expressed in pharyngeal gland cell g1; arrows point to the three nuclei of this large cell, arrowheads show two of the processes. (E)wrt-6::gfp expression in socket cells of the amphids (Amso) and one of the inner labial sensilla (Ilso) is shown. hyp5, an anterior hypodermal cell, is indicated. (F) L1 larva, dorsal view.wrt-5::gfp is seen in the seam cells, excretory cell (arrowhead), syncytial hypodermis, the six pharyngeointestinal valve cells (pi), rectal cells (re), and the pharynx corpus. (G)grd-1M::gfp is expressed in the rectal epithelial cells rect D, rect VL, and rectVR. (H) A posterior DA motorneuron expressing grd-7M::gfp; the nucleus (arrow) and axon (arrowheads) are shown. (I–K) grd-6C::gfp–lacZ is expressed in neurons of the head (I) and tail (K) as well as the bilateral HSN neurons (J, ventral view) of adult animals. The axon (arrowhead) belongs to the left nucleus (out of focus, toparrow); the right HSN nucleus is in focus (bottom arrow). The axon's twist is due to use of therol-6 coinjection marker. (L,M)grd-3C::gfp–lacZ (L) and grd-5C::gfp–lacZ(M) are expressed weakly in anterior seam cells of adults (arrows).
wrt-2::gfp is expressed in seam cells of 1.8-fold stage embryos to adults (Fig. 4B,C). At each seam, cell division GFP is initially present in both daughter cells and is subsequently lost in nonseam descendants. Additionally, most worms express GFP in hypodermal syncytia.
From the threefold embryo stage on, wrt-3::gfp is expressed in the trinucleate pharyngeal gland cell g1 (Fig. 4D). In addition, we see variable expression in seam cells and hypodermis, occasionally also in single muscle cells and distal tip cells of older larvae and adults. The predicted start codon of wrt-3 is only 250-bp downstream of the stop codon of a cGMP-gated cation channel exclusively expressed in neurons (Coburn 1996, C. Bargmann, pers. comm.). As expression patterns of these genes do not overlap, they do not seem to form an operon. Moreover, a 1-kb promoter fragment that does not include the channel gene is sufficient to express GFP in the described way, indicating that wrt-3 has its own promoter. In contrast, EST yk348a9 spans both the channel and wrt-3, suggesting a single transcript for the two genes. The exact nature of this unusual transcription complex is still a mystery.
wrt-5M::gfp expression starts at the beginning of morphogenesis in all seam cells and the excretory cell and remains until the adult stage. In larvae and adults, wrt-5 is also expressed strongly in the six cells of the pharyngeal-intestinal valve and the rectal valve (Fig. 4F). Phasmid socket cells and, as inwrt-6, sheath and/or socket cells of the anterior sensilla, express GFP in all postembryonic stages. Further, wrt-5M::gfpis expressed in adult animals in gonadal sheath cells, spermathecal sheath cells, and the uterus.
wrt-6M::gfp is expressed in four to seven sheath and socket cells of the anterior sensilla, judged from their neuron-like morphology and the position of their nuclei anterior to the pharynx metacorpus. Staining of amphid sensilla with DiI (Hedgecock et al. 1985) in transgenic worms show that amphid and inner labial socket cells express GFP. These cells are located on either side of the amphid process bundles (Fig. 4E). Some worms show additional expression in seam cells and hypodermal syncytia.
Three independently amplified promoter fragments for wrt-7 do not show any detectable GFP expression. By PCR of cDNAs we were only able to amplify a genomic fragment for wrt-7, presumably from contaminating genomic DNA in the cDNA preparation. Therefore,wrt-7 may be a pseudogene or expressed at very low levels only.
Expression Pattern of grd Genes
For grd-1, two different constructs, an amino-terminalgrd-1M::gfp and a grd-1C::gfp–lacZ reporter construct are expressed in three rectal cells of larvae to adult, possibly the rectal epithelial cells (Fig 4G). Reporter constructs forgrd-2M::gfp were not expressed. However, our recent computer analysis with the new genome data revealed that the methionine codon that we used for the grd-2 construct was very likely not the authentic initiation codon. The new grd-2 gene product has an extra 78 amino acids in front of the methionine residue that we used for the Met fusion construct (Fig. 3). This may explain the failure to see any expression.
The grd-3C::gfp–lacZ and grd-5C::gfp–lacZ reporter constructs gave lacZ staining in anterior and posterior seam cells in adult worms (Fig. 4L,M). grd-6M::gfp worms express GFP weakly in the head hypodermis, whereas grd-6C::gfp–lacZtransgenics show β-galactosidase signals in HSN neurons, several neurons in the head and tail (Fig. 4I–K). The additional expression seen in grd-6C::gfp–lacZ may require regulatory elements in introns that are not included in the start methionine fusion.grd-7M::gfp worms express GFP in three to four posterior DA motor neurons of the ventral nerve cord (Fig. 4H). None of thegrd-8 and grd-9 reporter constructs showed any GFP or β-galactosidase expression in transgenic animals.
DISCUSSION
Expression Patterns
Given the large number of wrt, grd, and grl genes, recent duplication events may have generated copies of genes that are redundant, may not have yet acquired a new function, or are not expressed anymore. For example, wrt-4, wrt-7, andwrt-8 are very similar, and wrt-7 does not seem to be expressed and may be a recent pseudogene. Other potential pseudogenes are listed in the online supplement table. But, overall, we estimate that <10%–20 % of the >50 genes in C. elegans are pseudogenes.
Our work has faced inherent problems when expression patterns of secreted molecules need to be examined. Methionine fusions have the advantage of avoiding the problem of secretion. However, some intronic regulatory elements may be omitted. More importantly, although we had some guidance for determining the start methionine by requiring a signal sequence for protein export, the precise start could not always be unambiguously determined, in particular for the grd genes. Thus, some constructs may have been fused to the wrong methionine, leading possibly to missed expression patterns.
Reporter constructs that are fused in the middle of the protein have the advantage of including introns with potential regulatory elements and the reading frame and exon can be properly determined because of sequence homology. Conversely, in such constructs, the GFP is secreted and the expression pattern cannot be evaluated when only diffuse fluorescence is seen in extracellular spaces, for example, in the body cavity. Sometimes no fluorescence at all is seen. Our use of vectors containing both gfp and lacZ did provide some benefits. For several genes (grd-3, grd-5, grd-6), we saw good expression using X-gal staining, in which no or only poor expression was seen with GFP.
The expression patterns observed for the wrt and grdgenes revealed diverse patterns, as we see expression in hypodermis, neurons, neuron-associated cells, and gland cells. Some genes are expressed in the same tissue and are of similar structure (e.g.,wrt-4, wrt-8, and wrt-1), so it is possible that these genes function in a combinatorial fashion or are partially redundant. Other genes are expressed in diverse sets of cells with no indication of overlapping expression. Overall, these genes may play a role in many different places, the only common denominator being that the cell types are mainly of ectodermal origin. Comparing the expression patterns of the C. elegans genes with those of thehh family is difficult at best, because the hh family genes function in a multitude of places (Hammerschmidt et al. 1997). Nevertheless, the diversity of wrt and grd expression patterns suggests that their functions could be equally diverse.
Evolution of Hog Genes
The Wart, Ground, and Ground-like domains display a high degree of sequence conservation in their cysteine patterns. However, several exceptions have been found, indicating that sudden changes are possible. For example, the B.m. wrt-6 homolog seems to have lost the second part of the Wart domain, and in several instances two cysteine residues have changed concomitantly. The latter observation suggests that particular cysteine residues may form disulfide bonds (Fig. 5A). The phylogenetic analysis of the Hog domains indicates that the different Hog domains are derived from each other as they are more related to each other than to inteins. Examination of the amino-terminal regions of the Wrt, Grd, Grl, M110, and Hh proteins revealed a putative conserved core region, C-FDφV-C (Fig. 5A).
As this motif is small, we tried to obtain additional evidence that would support the common origin of this motif. The sequence alignments of the Ground and Ground-like domains reveal conserved hydrophobic residues in spacings characteristic for amphipatic helixes within the first half of the motif. Secondary structure predictions for the multiple aligned Ground and the Ground-like domains confirm that the first half of the domains yield excellent scores for the formation of two α-helixes. The first helix starts at a conserved cysteine residue (Figs. 1C,D, and 5A). The second part of the domains is predicated to contain four, or perhaps three short β-strands that may form a β-sheet. The first β-strand after the α-helixes has a high score and it coincides with the conserved FDφV motif (Figs. 1C,D, and 5A). A possible topology based on allowed α–β sandwich structures (Fig. 9q in Orengo and Thornton 1993) is shown in Figure 5B.
The X-ray structure of the Hedge domain of Shh (Shh-N) has been determined (Hall et al. 1995). Shh-N contains two α-helixes; the first helix starts at a conserved cysteine residue and is followed by two β-strands and the second helix. After the helix, at the FDφV motif, another β-strand starts followed by a further β-strand that includes the last cysteine residue of the Hedge domain (Fig. 5A,B). The two cysteine residues are only 8.4 Å apart (Fig. 5B). Comparing the structure of Shh-N with the prediction of the Ground and Ground-like domains, we see some intriguing parallels: The first part of the core of the motif contains two α-helixes, the first of which starts at a cysteine residue; β-strands in the second part, the first of which starts at the FDφV motif after the second helix. Further, the proximity of the two cysteines in Shh-N lends support to the idea that the corresponding cysteines in the Ground and Ground-like domains may form a disulfide bond. There are also differences, such as the β-strands 3 and 4 between the two α-helixes of Shh-N (Fig. 5A,B). But overall, the structural and sequence similarities are striking and seem unlikely to be a chance coincidence.
In conclusion, the following lines of evidence indicate a monophyletic origin of hh, M110, wrt, grd, and grl genes: (1) Sequence and phylogenetic analysis of the Hog domains indicates that they are derived from a single ancestor. (2) The Hog domain is associated with amino-terminal domains that all have sequences for protein export. (3) Within all of these amino-terminal domains, a core sequence element C—FDφV–C can be found. (4) This core element coincides with the structural core of the Hedge domain. (5) Predictions and structure of the Ground, Ground-like and Hedge domains suggest similar structural features of these core elements.
We propose the following model for the evolution of the Hog genes (Fig.5C). At first, a single hog gene, nicknamedwedgehog, encoding a secreted amino-terminal domain existed. This ancestral gene may have arisen early in metazoan evolution from an intein, because this domain is more ancient as it occurs in prokaryotes. The wedgehog gene gave rise to hh and the ancestor of wrt and M110. Subsequently,M110 and wrt separated. wrt duplicated to give rise to wrt-10 and wrt-1. wrt-1 gave rise to additional copies of wrt genes, with and without Hog domains. One of the wrt genes gave rise to an ancestral grdgene. The Ground domain duplicated within the gene, and eventually, a duplicated copy of this gene gave rise to a gene cluster (grd-3/4 cluster) that lost the Hog domain. From this cluster, other Ground-only genes seem to be derived. Furthermore, somegrd gene gave rise to the grl genes. Overall, thegrd and grl gene proliferated and diversified rapidly.
When did hh and wrt/M110 duplicate and diverge from each other? The M110, Wart, Ground, and Ground-like domains have so far only been found in nematodes, although it might be difficult to detect sequence similarities outside of nematodes because of high sequence divergence. Further, no hh gene has been found in theC. elegans genome. Two different scenarios are possible: (1) If the genes duplicated before the separation of nematodes, thenC. elegans would have lost hh. (2) If the ancestralwrt/M110 is directly derived from the ancestralwedgehog or hh gene (depending on when in evolution nematodes separated from other phyla), then Wrt, M110, Grd, and Grl would be homologs of the vertebrate/fly Hh molecules.
We initially favored the first hypothesis. However, as sequencing projects proceed and no other Hog domains are found in other animal phyla, the second hypothesis becomes more likely. Thus, one has to entertain the notion that the wrt, grd, grl, and M110genes are the evolutionary homologs of the hh genes.
Possible Role of the New Gene Families
What is the function of the wrt, grd, and grl gene families? Recently, a Hh interactingprotein, Hip, has been identified (Chuang and McMahon 1999). However, database searches did not reveal any related molecule in C. elegans. A known target of Hh is Patched, which is part of the Hh-signaling cascade (Marigo et al. 1996). BLAST searches reveal >15 molecules with significant similarity to Patched in C. elegans, although some are more similar to Patched than the others (data not shown). The Patched molecule belongs to a group of integral membrane molecules with a sterol-sensing domain (SSD), which includes other proteins such as Niemann-Pick C and HMG CoA reductase (Beachy et al. 1997). When PSI–BLAST searches are performed with Patched, distantly related molecules identified are—among others—multidrug resistance molecules (data not shown). Generally, these molecules seem to sense or transport small organic compounds. How the C. elegans Patched-like molecules are related to the SSD molecules remains to be determined. But, given that the Hedge, Wart, Ground, and Ground-like domains evolved from each other, it is feasible that their targets coevolved. Thus, the diverse set of C. elegansPatched-like molecules could be among these targets. Perhaps one can think of the Wrt, Grd, and Grl proteins as molecules that bind and modulate the activity of Patched-like membrane molecules, reminiscent of the modulating activities of neuropeptides.
In Hh proteins, the Hog domain is responsible for anchoring the cholesterol adduct to the amino-terminal domain (Beachy et al. 1997). However, in C. elegans, the Wart and Ground domains can occur with or without associated Hog domain. It suggests that a fundamental function resides in the amino-terminal protein domain and does not depend on the addition of an adduct. Perhaps the major distinction between the Hog and non-Hog forms is that the former may be anchored to the membrane, whereas the latter are free to diffuse. One also needs to consider the efficiency of the cleavage reaction; the Hog domain could simply keep the amino-terminal Wart or Ground domain in an inactive state until autoproteolytic cleavage occurs.
METHODS
Sequence Analysis
C. elegans and C. briggsae database searches were performed at the Sanger Center (Cambridge, UK) and the Genome Sequencing Center (Washington University, St. Louis, MO) (http://www.sanger.ac.uk/Projects/Celegans/,http://genome.wustl.edu/gsc/) with their web implementation of BLAST (Altschul et al. 1990; W. Gish 1994–1997, unpubl.). Genomic organization of the gene structures was analyzed with GENEFINDER within the ACeDB database on a local workstation (Durbin and Thierry Mieg 1991). Some detailed sequence comparisons were performed with an interactive dot matrix program on the Macintosh (Bürglin 1998). Searches of Genbank were performed with Netblast at the National Center for Biotechnology Information (NCBI) (Altschul et al. 1990), or the Web BLAST and the PSI–BLAST servers at NCBI (Altschul et al. 1997). Several programs of the GCG package were used, such as Fetch, Gelassemble (for cDNA assembly), and Pileup in GCG (Devereux et al. 1984). Phylogenetic analysis was carried out by the program CLUSTALX on a Macintosh (Thompson et al. 1997), and trees were visualized NJPLOT by M. Gouy (http://biom3.univ-lyon1.fr/software/njplot.html). Aligned sequences were submitted to the PredictProtein server athttp://dodo.cpmc.columbia.edu/predictprotein/ (Rost 1996) for secondary structure prediction. Structures were viewed by RasMol2 for the Macintosh (ftp://ftp.dcs.ed.ac.uk/pub/rasmol/, Sayle and Milner-White 1995).
Cloning of Reporter Constructs and Analysis of Transgenic Animals
Reporter constructs were cloned by PCR amplification of cosmid orC. elegans genomic DNA. To reduce the possibility of PCR artifacts, we determined the expression of two PCR clones for each construct. Promoter fragments were 1.5–8 kb in size and, in most cases, covered the whole upstream sequence into the next upstream gene. The PCR fragments were cloned into either vector pPD114.108 (containing a nuclear localization sequence, NLS), pPD95.69, pPD95.70, pPD95.67, or pPD96.02, kindly provided by A. Fire, J. Ahnn, G. Seydoux, and S. Xu (Carnegie Institute of Washington, Baltimore, MD). For cloning, we either used restriction enzymes or a two-step PCR method we described earlier (Cassata et al. 1998). Primer sequences used are shown in the Online supplement.
Transgenic animals were obtained as described (Mello and Fire 1995;Mello et al. 1991). X-gal staining was performed as described (Fire 1992; Fire et al. 1990). Micrographs of worms were taken with a Nikon Microphot-FXA microscope with either DIC illumination for X-Gal-stained animals, or fluorescent illumination with a FITC B2A filter for GFP reporter constructs. Data were documented either on Ektachrome 400 slide film or directly captured with a cooled Sony DXC-950P color video camera (AVT Horn, Aalen, Germany) and a Scion CG-7 frame grabber (Scion, Inc., Frederick, MD) running on an Apple Power Macintosh computer.
cDNA Cloning
Partial cDNAs for wrt-5 and wrt-6 were isolated by PCR screening of a cDNA library (Okkema and Fire 1994) with gene-specific primers alone (wrt5ko5, GCATCCGACCACCATGTGCTCC; wrt5ko1, AAAAGGCAACCAATCGCTTAGCAACC), or in combination with vector specific primers (wrt6ko4, CATTGCTGTCTGTGGTGACGTGAATGC; wrt6ko3, TCATCCATTTTGACTTCTTTCGCGGCG; GT2, CGGATCCGTAGCGACCGGCGCTCAGCT). A partial cDNA for wrt-3 was isolated by PCR on a cDNA preparation from him-5 mixed stage animals with the primers wrt3ko6 (ATGTTGTACCACGTGGAAATGTTC) and wrt3ko7 (CGAATATCTTTTATTACATCAAATTTTACC). The cDNAs were sequenced and submitted to Genbank: wrt-3 (w3c9), AF139522; wrt-5(w5c2), AF139520; wrt-6 (w6c28), AF139521.
GenBank accession numbers for cDNA clones are given as follows:wrt-1, U61235 (Porter et al. 1996a); wrt-2, yk145f9,C12010, and C10408; wrt-4, cm02c8 (complete cds), and U61236;wrt-8, cm20f10, M89293, and U61237 (complete cds);grd-1, yk42d3, D34193, D37242, and U61288; grd-2, yk87f2, D71849, and D74573; grd-3, yk66e9, D65712, and D69245;grd-5, yk256h6, C40963, and C30673; grd-6, yk68a8, D69286.
Acknowledgments
We thank Andy Fire, Joohong Ahnn, Geraldine Seydoux, Siqun Xu, Peter Okkema, Giuseppe Cassata, and Yuji Kohara for sharing vectors, cDNAs, and libraries. We thank Arsène Gschwind, Rainer Pöhlmann, and Richard Durbin for help with computer problems. We thank Cara Coburn, Cori Bargmann, Geraldine Seydoux, Thomas Marty, Jay Groppe, and laboratory members for helpful discussions and sharing unpublished results. We are grateful to the Genome Sequencing Center for communication of DNA sequence data prior to publication. This work and G.A. are supported by a grant from the Wolfermann-Naegeli Stiftung. T.R.B. is supported by grant NF. 3130-038786.93. H.K. is a recipient of a grant from the Sandoz Stiftung and a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad. Additional support has been provided by the Swiss National Science Foundation and the University of Basel, Kanton Basel-Stadt.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵1 Corresponding author.
-
E-MAIL burglin{at}ubaclu.unibas.ch; FAX 41 61 267 2078.
-
- Received May 12, 1999.
- Accepted August 3, 1999.
- Cold Spring Harbor Laboratory Press