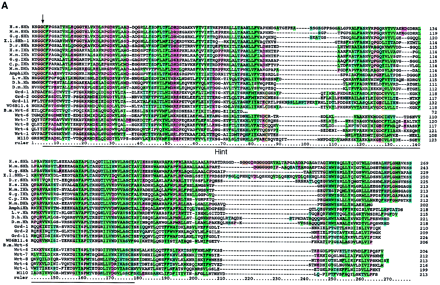
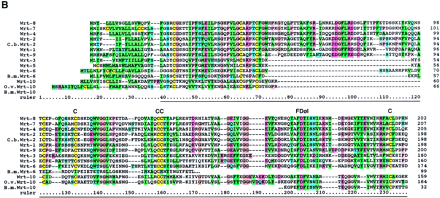

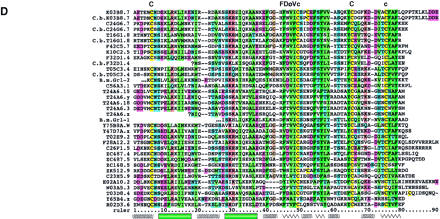
Sequence alignments of the Hog/Hint domains and associated motifs. The color coding of the residues is slightly modified from the default settings in ClustalX. In particular, small hydrophobic residues are green, large hydrophobic residues are dark green, and cysteine residues are yellow in all positions. In B–D the residues of the core motif (C-FDφV-C, see text) are shown above the alignments; the line under the first Cys in C indicates a variable position, small c in D indicates additional conserved residues. (A) Alignment of Hh Hog domains and the nematode Hog domains. (Arrow) The site of autocatalytic cleavage. A line marks the definition of the Hint domain. The whole alignment was used for the neighbor joining (NJ) analysis in Fig. 2A. Sequences used were taken from Bürglin (1996) with the addition of sea urchin (L.v.Hh,Lytechinus variegatus, GenBank accession no. AF059606), Amphioxus (AmphiHh, Y13858), newt (C.p.Hh, Cynops pyrrhogaster, D63339), and Drosophila hydeii (D.h.Hh, AAB34104). Other species codes are: (H.s.) human; (M.m) mouse; (G.g.) chicken; (D.r.) zebrafish; (X.l.) Xenopus laevis; (D.m.)Drosophila melanogaster. (B) Alignment of the Wart domains. The region use for the NJ analysis is marked by a line. (C) Alignment of the Ground domains. (rp) Repeat number. Beneath the secondary structure, prediction for the alignments is indicated. Squiggles mark loop regions, green bars α-helixes, and zigzag lines β-strands. Y102A5c.34 needs two frameshifts near the amino terminus—marked with a small x—to line up all the residues; Y69A2A.x has also some problems with extending the ORF over the conserved region, although this YAC sequence is still unfinished. (D) Alignment of the Ground-like domains. Secondary structure predictions as for C. The reading frame for T24A6.3 was changed by hand to get a good methionine with a signal sequence. T24A6.x seems to contain only the last two-thirds of the Ground-like domain. The sequence similarity of C.b.F32D1.4 extends over only one exon, with no other similarities in the flanking regions to be found.C.b.F32D1.4 might be a recent pseudogene, suggesting thatF32D1.4 may not be highly conserved in evolution.











