Microsatellite Marker Content Mapping of 12 Candidate Genes for Obesity: Assembly of Seven Obesity Screening Panels for Automated Genotyping
Abstract
Twin studies, adoption studies, and studies of familial aggregation indicate that obesity has a genetic component. Whereas, the genetic factors predisposing to obesity have been elucidated for several rare syndromes, the factors responsible for obesity in the general population have remained elusive. Genetic studies of complex traits are often accelerated by the use of candidate genes. To facilitate genetic studies of human obesity, seven multiplex panels of candidate genes for obesity that are suitable for fluorescent genotyping have been assembled. The multiplex panels are composed of 66 microsatellite markers linked tightly to 16 human gene products that are of potential importance in the control of body weight or linked to syndromic forms of obesity. As part of these efforts 12 previously cloned genes have been placed on the human physical map. In addition the chromosomal location of three of these genes, ART,NYP Y6R, and PPARγ, are reported for the first time. These resources will be of use in studies to identify the genetic factors responsible for human obesity. [Figures are available at http://www.genome.org.]
Obesity is highly prevalent in Western society, affecting approximately one-third of the United States population and 20% of Europeans. An increased body weight predisposes to diabetes, heart disease, and hypertension and is a major cause of morbidity and mortality. Several lines of evidence suggest that genetic factors play an important role in the development of obesity. However, despite recent progress in human genetics, it has proven difficult to localize human obesity genes using family studies. Thus, whereas the genes for several syndromes that include obesity as a feature have been localized, linkage to an obesity gene in the general population has proven to be exceedingly difficult. The identification of obesity genes will likely require the genotyping of large numbers of affected individuals with a dense array of genetic markers. Such genetic studies are often facilitated by the inclusion of genetic markers that represent candidate genes. Candidate genes for human obesity would include the human homologs of rodent obesity genes, as well as other molecules that have been suggested to play a role in regulating weight. A partial list of these genes includes agouti-related transcript (ART), neuropeptide Y (NPY) and its receptors NPY Y5R and Y6R,pro-opiomelanocortin (POMC), uncoupling protein 2 (UCP2), and the melanocortin-4 receptor(MC4R) (Erickson et al. 1996; Gerald et al. 1996; Weinberg et al. 1996; Comuzzie et al. 1997; Fleury et al. 1997; Huszar et al. 1997;Shutter et al. 1997).
To facilitate the use of these candidate genes as markers in genetic studies of obesity, each has been localized on the human genetic and physical maps by screening of a human yeast artificial chromosome (YAC) library and radiation-hybrid (RH) panels. We report the chromosomal location of several of these genes the first time. In addition, microsatellites in the vicinity of each of these genes, (i.e., within a YAC contig) have been compiled into seven multiplex panels suitable for automated genotyping. The use of these panels is likely to facilitate the analysis of these loci in family studies of human obesity.
RESULTS AND DISCUSSION
Candidate Gene Microsatellite Content Mapping
The identification of human obesity genes has proven difficult partly because of the fact that human obesity is genetically polygenic and heterogenous. The importance of identifying obesity genes has been amplified by the successful cloning of each of the five single-gene mutations that cause obesity in rodents. The isolation of the mouseob and db genes have identified leptin and its receptor as components of a negative-feedback loop regulating the size of the adipose tissue mass (Zhang et al. 1994; Tartaglia et al. 1995;Lee et al. 1996). The agouti gene modulates melanocortin signaling in the hypothalamus and inhibits the anorexogenic effect of melanocyte-stimulating factor (MSH) in this brain region (Wilson et al. 1995; Fan et al. 1997). The fat and tubby mutations also appear to alter the neural circuits that regulate weight. Thefat gene encodes carboxypeptide E (CPE), an enzyme involved in neuropeptide processing (Naggert et al. 1995). Thetubby (tub) gene is expressed in the paraventricular nucleus of the hypothalamus, a brain region known to play a role in the regulation of body weight (Kleyn et al. 1996; Noben-Trauth et al. 1996).
In aggregate, these data have suggested that body fat is controlled by a lipostat mechanism in which leptin is the afferent signal and the hypothalamus serves as an integrator, thus activating an output loop that modulates feeding behavior, energy expenditure, and fat and glucose metabolism (Friedman 1997). It is likely that the genes that predispose to obesity will function within this system and encode factors that modulate leptin secretion, regulate the hypothalamic integration of nutritional signals, or play a role in the effector limb of this system. Several such candidates have been identified recently including ART, apolipoprotein J (APOJ),β-3 adernergic receptor(β-3AR), CPE, MC4R, NPY Y5R, NPY Y6R, prohormone convertase 1 (PC 1), POMC, peroxisome proliferator-activated receptor γ (PPAR γ), tub, and UCP2 (Tontonoz et al. 1994; Naggert et al. 1995; Clement et al. 1996; Gerald et al. 1996; Kleyn et al. 1996; Noben-Trauth et al. 1996; Weinberg et al. 1996; Comuzzie et al. 1997; Fleury et al. 1997;Huszar et al. 1997; Jackson and Li 1997; Shutter et al. 1997; R. Lallone, pers. comm.).
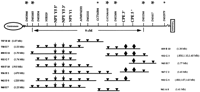
Each of these candidates was localized on the human genetic and physical maps by screening a radiation hybrid (RH) panel and/or a human YAC library. These genes were localized in YAC contigs and the contigs were assayed for their STS content. One region of human chromosome 4 contains the NPY Y1R and Y5R genes as well as theCPE gene (Fig. 1). (For a complete listing of genes, see Fig. 1 in its entirety at http://www.genome.org.) In this case, STS-content mapping made possible the selection of microsatellites that distinguish the NPY Y1/Y5R genes fromCPE in linkage studies. The analysis of each of these candidate genes is described in turn.
STS content maps. The chromosome 4 subregion containing the NPY Y1R, NPY Y5R, and CPE genes is shown as an example of those generated in this study. The orientation of theCPE gene is indicated by a 3′ or 5′ following theCPE gene symbol and runs left to right. The orientation of the NPY Y5R and NPY Y1R genes could not be determined by the use of 3′ and 5′ primer sets relative to each other: YACs are shown as lines, the lengths of which reflect the number of included STSs and not actual size. Total size of individual YACs is listed next to the YAC name. The physical distances between adjacent STSs have not been determined directly. Inverted triangles indicate that the YAC was positive for the indicated STS: stars indicate that the YAC was positive for a particular candidate primer set; and diamonds indicate that the YAC was positive for a particular CPE primer set. Genetic distances (in cM) are indicated between selected STSs. Only YACs that were positive with more than one STS are shown. Asterisks above a STS name indicate a polymorphic marker with larger symbols denoting inclusion in a screening panel. Physical (STS content) maps for the following genes are available on the Genome Research web site http://www.genome.org: ART, APOJ,β-3AR, MC4R, NPY Y6R, PC 1, POMC, PPARγ, tub, andUCP2.
ART
Oligonucleotide primers based on both the 5′ and 3′ sequences of the human ART cDNA sequence (GenBank accession no. U88063) were used to screen a CEPH B YAC library and the GeneBridge 4 (Research Genetics) human–rodent somatic RH panel by PCR. The RH screen placed ART at 2.94 cR from STS WI-5594, which is 299.70 cR from the top of human chromosome 16. Six YACs were selected from the contigs WC16.5, 16.6, and 16.7 of the Whitehead anchored map for human chromosome 16, and 5 of 6 of the candidate YACs were confirmed by PCR as containing the ART sequence (Tsaur et al. 1995). These YACs 767 C 6, 782 G 9, 837 F 9, 877 C 6, and 898 G 5 are part of contigs WC16.5, 16.6, and 16.7 of the Whitehead anchored map and are localized to the 16q21–22.1 region of chromosome 16 (Hudson et al. 1995). The alignment of the ART YACs relative to the known map positions of the microsatellite was elucidated from microsatellite mapping and arranged in a centromeric-to-telomeric manner as follows: 837 F 9, 898 G 5, 877 C 6, 767 C 6, and 782 G 9. Polymorphic microsatellites D16S3019 and D16S496 are contained on the smallest ART YAC 782 G 9 and are therefore within 555 kb of ART. Microsatellites D16S400 andD16S302 are contained at the border on the ART YAC contig and are also polymorphic. The four microsatellites are contained within a 3-cm distance on the Whitehead chromosome 16 anchored contig map (Hudson et al. 1995).
APOJ
Oligonucleotide primers based on the human APOJ cDNA sequence (GenBank accession no. J02908) and corresponding to the 3′ untranslated region of the human APOJ cDNA were used to screen a CEPH B YAC library and the GeneBridge 4 human–rodent somatic RH panel by PCR. The RH screen placed APOJ at 3.46 cR from STS WI-1172 which is 111.17 cR from the top of human chromosome 8. Seven YACs were selected from the contig WC 8.4 of the Whitehead anchored map for human chromosome 8, and 4 of 7 of the candidate YACs containedAPOJ (Tsaur et al. 1995). The APOJ gene containing YAC contig overlaps in a centromeric-to-telomeric manner as follows: 755 B 3, 771 G 9, 791 B 9, and 810 D 6. Three of these YACs, 755 B 3, 771 G 9, and 791 B 9, are part of contig WC8.4 of the Whitehead anchored map and are localized to the 8p21.3–q11.1 region of chromosome 8 (Hudson et al. 1995). Polymorphic microsatellitesD8S137 and D8S1809 are contained on the smallestAPOJ YAC 810 D 6 and are therefore within 750 kb ofAPOJ. In addition, the polymorphic tetranucleotide repeat STS GAAT13A04 borders the contig (map available as supplementary information on www.genome.org). D8S137 andD8S1809 are contained within a 4-cm distance on the Whitehead Chromosome 8 anchored contig map (Hudson et al. 1995).
β-3AR
Oligonucleotide primers based on the humanβ-3AR (β3AR) cDNA sequence (GenBank accession no. X72861) and corresponding to the 3′ region of the human β3AR cDNA were used to screen a CEPH B YAC library by PCR. Four β3ARgenes containing YAC clones were identified in the screen mapping to human chromosome 8. The contig formed by the fourβ3AR genes containing YACs is as follows: 776 C 2, 807 A 2, 770 C 2, and 841 B 7. The β3ARreceptor YAC contig overlaps in a telomeric-to-centromeric manner. These YACs are part of contig WC 8.5 of the Whitehead anchored map and are localized to the 8p12–p11.2 region of chromosome 8 (Hudson et al. 1995). Polymorphic microsatellite D8S1791 is contained on the smallest β3AR YAC 841 B 7 and is therefore within 840 kb of β3AR. In addition, the polymorphic tetranucleotide repeat microsatellite GATA101H09borders the β3AR YAC contig (map available as supplementary information on www.genome.org). Theβ3AR contig is contained within a 2-cm distance on the Whitehead chromosome 8 anchored contig map (Hudson et al. 1995).
CPE
Oligonucleotide primers based on the human CPE cDNA sequence (GenBank accession no. X51405) and corresponding to the 5′-untranslated region of the human CPE cDNA were used to screen a CEPH B YAC library by PCR. Five YAC clones were identified in the screening as containing the CPE sequence. The contig formed by the five CPE-gene-containing YACs is, as elucidated from microsatellite mapping, as follows: 932 G 5, 859 B 10, 932 G 3, 967 C 2, and 945 H 7. The CPE YAC contig overlaps in a centromeric-to-telomeric manner. These YACs are part of contig WC 4.6 of the Whitehead anchored map and are localized to the 4pter–qter region of chromosome 4 (Hudson et al. 1995). Tetranucleotide repeat microsatellites D4S2406 and GATA86A05 are contained in three of the informative YACs, including the smallest single-sized YAC 859 B10, and are therefore within 1.38 Mb of CPE (map available as supplementary information on www.genome.org). Polymorphic markers D4S1566 and D4S417 are also contained within the CPE YAC contig.
MC4R
Oligonucleotide primers based on the human MC4R cDNA sequence (GenBank accession no. S77415) and corresponding to the 3′ region of the human MC4R cDNA were used to screen a CEPH B YAC library by PCR. Nine MC4R-gene-containing YAC clones were identified in the screen mapping to human chromosome 18. The chromosomal location was confirmed by RH mapping with the MC4Rprimer set used in the YAC screening. The contig formed by the nineMC4R-gene-containing YACs is as follows: 933 F 9, 938 E 1, 943 B 8, 738 E 2, 772 F 6, 924 G 12, 883 A 7, 883 B 7, and 924 F 12. TheMC4R YAC contig overlaps in a telomeric-to-centromeric manner. These YACs are part of contig WC 18.4 of the Whitehead anchored map and are localized to the 18q21.32 region of human chromosome 18 (Hudson et al. 1995). The polymorphic tetranucleotide repeat microsatelliteGATA89B12 is contained in all nine of the informative YACs, including the smallest YAC 883 A 7, and is therefore within 870 kb ofMC4R. Polymorphic microsatellites D18S1357(tetranucleotide repeat) and/or D18S64 each are in five of nine of the informative YACs (map available as supplementary information on www.genome.org). The three microsatellites are contained within a 6-cm distance on the Whitehead chromosome 18 anchored contig map (Hudson et al. 1995).
NPY Y5R
Oligonucleotide primers based on the human NPY Y5R cDNA sequence (GenBank accession no. U56079) and corresponding to the 3′ region of the human NPY Y5R cDNA were used to screen a CEPH B YAC library by PCR. Seven NPY Y5R-gene-containing YAC clones were identified in the screen mapping to human chromosome 4. The contig formed by the seven NPY Y5R-gene-containing YACs is as follows: 890 E 11, 968 E 7, 758 E 7, 932 G 7, 935 F 10, 962 D 5, and 956 H 1. The NPY Y5R YAC contig overlaps in a centromeric-to-telomeric manner. These YACs are part of contig WC 4.6 of the Whitehead anchored map and are localized to the 4q31–32 region of human chromosome 4 (Hudson et al. 1995; Herzog 1997). All seven YACs containing the NPY Y5R gene contain nonpolymorphic microsatellite marker WI7198, which corresponds to theNPY Y1R gene as reported by the Whitehead map (Hudson et al. 1995). The polymorphic dinucleotide repeat microsatelliteD4S1603 is contained in two of the informative YACs, including YAC 968 E 7 and is therefore within 1.23 Mb of NPY Y5R.Polymorphic microsatellite D4S3046 borders on the centromeric end of the NPY Y5R YAC contig (map available as supplementary information on www.genome.org). The NPY Y5R-contig, theNPY Y1R gene (which is said to be in the opposite orientation of NPY Y5R) and the CPE locus are all found in WC 4.6 of the Whitehead map and are thus localized within the same 9-cm region of chromosome 4 (Gerald et al. 1996).
NPY Y6R
Oligonucleotide primers based on the human NPY Y6R cDNA sequence (GenBank accession no. D86519) and corresponding to the 3′ untranslated region of the human NPY Y6R-cDNA were used to screen a CEPH B YAC library and the GeneBridge 4 human–rodent somatic RH panel by PCR. The RH screen placed NPY Y6R at 4.19 cR from STS AFM350YB1, which is 431.25 cR from the top of human chromosome 5. Four YACs were found to contain the NPY Y6R-sequence and were from the contig WC 5.10 of the Whitehead anchored map for human chromosome 5. The contig formed by the four NPY Y6R-gene-containing YACs is as follows: 848 D 1, 880 G 9, 829 E 1, and 784 B 4. The NPY Y6R YAC contig overlaps in a centromeric-to-telomeric manner and is oriented 3′ to 5′ relative to the centromere. Polymorphic microsatellite D5S816, a tetranucleotide repeat, is contained on the smallest NPY Y6RYAC 880 G 9 and is therefore within 890 kb of NPY Y6R.Additional polymorphic microsatellite markers D5S1983,D5S414, and D5S500 are contained by each of the other three YACs within the NPY Y6R-contig (map available as supplementary information on www.genome.org). All four microsatellites are contained within a 1-cm distance on the Whitehead chromosome 5 anchored contig map (Hudson et al. 1995).
PC 1
Oligonucleotide primers based on the human PC 1 cDNA sequence (GenBank accession no. X64810) and corresponding to the 3′ region of the human PC 1 cDNA were used to screen a CEPH B YAC library by PCR. Seven PC 1-sequence-containing YAC clones were identified in the screen mapping to human chromosome 5. The contig formed by the seven PC 1-gene-containing YACs is as follows: 815 B 5, 872 F 12, 845 F 12, 947 C 8, 955 E 6, 960 D 12, and 770 A 10. The PC 1 YAC contig overlaps in a centromeric-to-telomeric manner, and the PC 1 gene is oriented 5′ to 3′ relative to the centromere. These YACs are part of contig WC 5.7 of the Whitehead anchored map and are localized to human chromosome 5 (Hudson et al. 1995). The polymorphic tetranucleotide repeat microsatelliteGATA3H06 and dinucleotide repeat D5S484 are contained in four of seven of the informative YACs, including YAC 770 A 10, and are therefore within 420 kb of PC 1. In addition, polymorphic microsatellite D5S644 is contained in six of seven YACs within the PC 1 contig. All three microsatellites are contained within a 1-cm distance on the Whitehead chromosome 5 anchored contig map (Hudson et al. 1995). The PC 1 YAC contig contains nonpolymorphic STS WI7819, which corresponds to thecalpastatin gene, mapping to position 5q15–q21 of chromosome 5. This positions the PC 1 gene to this chromosomal location (map available as supplementary information on www.genome.org).
POMC
Oligonucleotide primers based on the human POMC cDNA sequence (GenBank accession no. S59424 and M38297) and corresponding to the 3′ and 5′ region of the human POMC cDNA were used to screen a CEPH B YAC library by PCR. FivePOMC-gene-containing YAC clones were identified in the screen mapping to human chromosome 2. The contig formed by the fivePOMC-gene-containing YACs is as follows: 744 H 7, 887 D 8, 903 D 10, 931 E 4, and 713 G 9. The POMC YAC contig overlaps in a telomeric-to-centromeric manner. These YACs are part of contig WC 2.2 of the Whitehead anchored map and are localized to 2p24–p21 region of human chromosome 2 (Hudson et al. 1995). The polymorphic microsatellites D2S144 and D2S171 are contained in all five of the informative YACs, including YAC 931 E 4, and are therefore within 370 kb of POMC. In addition, polymorphic microsatellites D2S2168 and D2S2170 are contained within the POMC contig (map available as supplementary information on www.genome.org). All four microsatellites are contained within a 3-cm distance on the Whitehead chromosome 2 anchored contig map (Hudson et al. 1995).
PPARγ
Oligonucleotide primers based on the humanPPARγ cDNA sequence (GenBank accession no.L40904) and corresponding to the 3′ region of the humanPPARγ cDNA were used to screen a CEPH B YAC library by PCR. Four YAC clones were identified in the screen mapping to human chromosome 3. Two of four of the candidate YACs were confirmed by PCR as containing the PPARγ sequence (Tsaur et al. 1995). The contig formed by the twoPPARγ-gene-containing YACs is as follows: 754 D 7 and 897 A 6. The PPARγ contig overlaps in a centromeric-to-telomeric manner. These two YACs are part of contig WC 3.3 of the Whitehead anchored map and are localized to the 3pter–qter region of human chromosome 3 (Hudson et al. 1995). BothPPARγ YACs contain the polymorphic microsatellite markers 32 cm from the top of chromosome 3 as reported by the Whitehead map (Hudson et al. 1995).
The polymorphic microsatellite D3S3701 is contained in both of the informative YACs, including YAC 754 D 7, and is therefore within 200 kb of PPARγ. ThePPARγ YAC contig is contained within a 2-cm region of chromosome 3, whose border is defined by the polymorphic microsatellite D3S3610 (map available as supplementary information on www.genome.org).
tub
Oligonucleotide primers based on the human tub homolog cDNA sequence (GenBank accession no. U54644) and corresponding to the 3′ region of the human tub cDNA were used to screen a CEPH B YAC library by PCR. Six tub-gene-containing YAC clones were identified in the screen mapping to human chromosome 11. The contig formed by the six tub-gene-containing YACs is as follows: 716 F 4, 721 E 11, 817 A 7, 960 F 10, 954 F 4, and 927 B 2. Thetub YAC contig overlaps in a telomeric-to-centromeric manner. These YACs are part of contig WC 11.0 of the Whitehead anchored map and are localized to the 11p15.4–15.1 region of human chromosome 11 (Hudson et al. 1995). The polymorphic dinucleotide repeat microsatellites D11S932 and D11S1331 are contained in five of six of the informative YACs, including YAC 716 F 4, and are therefore within 300 kb of tub (map available as supplementary information on www.genome.org). In addition, polymorphic microsatelliteD11S909 is contained within the tub contig. Microsatellites D11S1331 and D11S909 define the borders of a 2-cm region on the Whitehead chromosome 11 anchored contig map that contains the tub contig (Hudson et al. 1995).
UCP2
Oligonucleotide primers based on the human UCP2 cDNA sequence (GenBank accession no. U76367) and corresponding to the 3′ region of the human UCP2 cDNA were used to screen a CEPH B YAC library by PCR. Four UCP2-gene-containing YAC clones were identified in the screen mapping to human chromosome 11. The contig formed by the four UCP2-gene-containing YACs is as follows: 744 E 7, 870 E 8, 943 H 2, and 858 H 9. The UCP2 YAC contig overlaps in a centromeric-to-telomeric manner. These YACs are part of contig WC 11.9 of the Whitehead anchored map and are localized to 11q13–q23 region of human chromosome 11 (Hudson et al. 1995). The polymorphic microsatellite D11S4207 is contained in all four of the informative YACs, including YAC 744 E 7, and is therefore within 1.04 Mb of UCP2. In addition, polymorphic tetranucleotide repeat microsatellite D11S2371 and dinucleotide repeatD11S916 are contained within the UCP2 contig (map available as supplementary information on www.genome.org). All three microsatellites are contained within an ∼3 cm distance on the Whitehead chromosome 11 anchored contig map (Hudson et al. 1995).
Candidate Gene and Syndrome-Screening Panels
The localization of these obesity candidate genes on a physical map allowed the compilation of a list of polymorphic microsatellite markers within each of the aforementioned contigs. In addition to the 12 candidate genes described above, microsatellites in proximity to four previously mapped genes, agouti, leptin, theleptin receptor, and NPY were also included (Stirling et al. 1995; Wilson et al. 1995; Clement et al. 1996; Winick et al. 1996; Bray, pers. comm.).
These markers, together with markers linked previously to candidates for syndromic or other forms of human obesity, were assembled into multiplex panels suitable for automated genotyping. Genotyping technologies utilizing the fluorescent dyes FAM, TET, and HEX allow for the pooling of separate PCR reactions with similarly sized products prior to electrophoresis in a single lane of a polyacrylamide gel (Schwengel et al. 1994; S. Diehl, J. Siegle, G.A. Buck, T.R. Reynolds, and J.L. Weber, unpubl.). In this manner, 9–12 separate genotypes can be analyzed at once (depending on the range of the expected product sizes for the microsatellites).
Seven screening panels (Table 1) comprised of 66 polymorphic microsatellites from 16 genes were assembled to take full advantage of the attributes of the fluorescent genotyping technologies. Two of the seven panels (Table 1, panels 6 and 7) are comprised of microsatellites markers for human syndromes in which obesity is a salient feature of the disease phenotype. These syndromes include Bardet-Biedl 1-4, Schinzel syndrome, Cohen’s syndrome, Angelman syndrome, and Prader–Willi syndrome (Kwitek-Black et al. 1993; Leppert et al. 1994; Sheffield et al. 1994; Tahvanainen et al. 1994; Bamshad et al. 1995; Carmi et al. 1995; Robinson and Lalande 1995; Kishino et al. 1997). The panels were each composed of 9–11 microsatellites and were organized to provide maximal separation between similarly labeled PCR products, as different populations often exhibit vastly different size ranges for a given microsatellite. The panels were constructed to allow for differences in the size ranges among diverse populations.
Candidate Gene Screening Panels
In selecting polymorphic STS markers for the construction of marker panels for candidate genes and obesity syndromes, additional criteria were also considered. Preferences was given to those STS with high maximum heterozygosity, and when possible, more than one STS from each contig was included. Multiple STS per gene allow potentially the delineation of segment-linkage disequlibrium in a population. Each of the STS was derived from a YAC (or in a YAC contig) containing the candidate gene. When possible, STS markers for both 5′ and 3′ of the candidate were included. Each STS is also anchored on the Whitehead STS-based map. When possible, tri- and tetranucleotide repeats were chosen to facilitate allele calling. Alternatively, robust dicucleotide repeats were selected. In the selection of markers from previously mapped candidate genes and obesity syndromes, the markers that best fit the criteria above were chosen.
The seven panels were constructed with the objective of combining the speed and accuracy of automated fluorescent genotyping while targeting specific obesity-related candidate genes. Although the use of these panels should not preclude a genome scan, their use could accelerate efforts to elucidate the genetic basis of human obesity. As new genes are discovered they can be mapped similarly and included in the candidate-gene screening panels. The composition of these panels is shown (Table 1).
METHODS
Candidate genes were chosen because of their roles in rodent obesity models or their implied roles in human obesity disorders. Oligonucleotide primers were generated for the 5′ and 3′ regions of obesity candidate genes against their published GenBank cDNA or genomic sequences. The candidate genes are as follows: ART(GenBank accession no. U88063), APOJ (GenBank accession no.J02908), β-3AR (GenBank accession no. X72861),CPE (GenBank accession no. X51405), MC4R (GenBank accession no. S77415), NPY Y5R (GenBank accession no. U56079),NPY Y6R (GenBank accession no. D86519),PPARγ (GenBank accession no. L40904),POMC (GenBank accession nos. S59424 and M38297), tub(GenBank accession no. U54644), and UCP2 (GenBank accession no. U76367).
Oligonucleotide primers for ART are as follows: ART66F (5′-TAATCGGCTCCTGGAAACCT-3′), ART 278R (5′-CTCTGCCTCCGGGATTCTTG-3′), ART 619F (5′-ACGTGCTACTGCCGCTTCTT-3′), and ART 745R (5′-GTTGGTCCCATCCTTTATTC-3′). Oligonucleotide primers forAPOJ are as follows: APOJ 1025F (5′-GGCTTCCCACACTTCTGACT-3′) and APOJ 1333R (5′-GGGAGAGGCTGGGCGGAGTT-3′). Oligonucleotide primers forβ-3AR are as follows:β-3AR 717F (5′-CCAGTGGGCTGCCAGGGG-3′) andβ-3AR 947R (5′-GCCAGTGGCGCCCAACGG-3′). Oligonucleotide primers forCPE are as follows: CPE 39F (5′-GCCTCGCAGTGGTTTCTCCT-3′), CPE 241R (5′-GCGGGCGGCCTCTTTTGTCT-3′), CPE 2104F (5′-ATGAATGCTATTGAAAAGGT-3′), and CPE 2363R (5′-ACCAAGAGAAAACCCTAAAC-3′). Oligonucleotide primers forMC4R are as follows: MC4R 52F (5′-ATGGCATGGCAGCTTCAAGG-3′), MC4R 255R (5′-GCCTGCTGTGAGTAAATGTC-3′), MC4R 1434F (5′-TTTTTCACTCTTACCCTACC-3′), and MC4R 1622R (5′-AATCCACAGTGCCTACAACC-3′). Oligonucleotide primers forNPY Y5R are as follows: NPY Y5R 92F (5′-CTTGCCACAGAGAATAATAC-3′), NPY Y5R 220R (5′-TAAGTAGATTCCCCATAAAG-3′), NPY Y5R 1198F (5′-GATGCCACTACACCTTTTCC-3′), and NPY Y5R 1368R (5′-ATGAAGACAGTGTATAAGGG-3′). Oligonucleotide primers forNPY Y6R are as follows: NPY Y6R 48F (5′-GCTGTTACATTCCTTGCCTC-3′), NPY Y6R 263R (5′-ATGTGAATGACTTGAGCGTG-3′), NPY Y6R 1732F (5′-CATACCACCCCTTTTCTCTT-3′), and NPY Y6R 1912R (5′-GTGTTTTTACTAGGCATATC-3′). Oligonucleotide primers forPPARγ are as follows:PPARγ 250F (5′-AGACCACTCCCACTCCTTTG-3′), PPARγ359R (5′-AGGTCATACTTGTAATCTGC-3′),PPARγ 1480F (5′-GCTCCAGAAAATGACAGACC-3′), and PPARγ1630R (5′-TGGAAGAAGGGAAATGTTGG-3′). Oligonucleotide primers forPOMC are as follows: POMC 12F (5′-CAAACAATGGGGAAATCGGA-3′), POMC 268R (5′-CGCTGGAAAGGGGCTGGAAT-3′), POMC 617F (5′-TGGCGGCCGAGAAGAAGGAC-3′), and POMC 746R (5′-CTTGATGATGGCGTTTTTGA-3′). Oligonucleotide primers fortub are as follows: tub 1741F (5′-GCCCTGCCTATCCTCTGTAT-3′) and tub 1923R (5′-AGGGTGGGAGTGTGTGTTGA-3′). Oligonucleotide primers forUCP2 are as follows: UCP2 21F (5′-AGATGTGCCCCCTACTGCCA-3′), UCP2 125R (5′-CTGACTTTCTCCTTGGATCT-3′), UCP2 709F (5′-GTGGTCAAGACGAGATACAT-3′), and UCP2 825R (5′-AACCCAAGCGGAGAAAGGAG-3′).
Oligonucleotide primers based on both the 5′ and 3′ sequences of candidate gene cDNA sequences were used to screen a CEPH B YAC library and the GeneBridge 4 (Research Genetics) human–rodent somatic RH panel by PCR. Candidate-gene-positive YACs were confirmed by PCR as containing the candidate gene of interest (Tsaur et al. 1995). YAC contigs were aligned for the various candidate genes relative to the known map positions of the microsatellites contained within the contigs as elucidated from microsatellite mapping. YAC contigs were arranged in a telomeric-to-centromeric manner when in a chromosomal p(short arm) region or in a centromeric-to-telomeric manner when in a chromosomal q (long arm) region.
The PCR was performed in a 25-ml (YAC) or 10-ml (RH) reaction mixture containing 3 ml blockpool mega YAC DNA or 25 ng genomic hybrid DNA in conditions described previously using 1.5 units of Taqpolymerase (Boehringer Mannheim, Germany) (Winick et al. 1996). DNA samples were subjected to 35 cycles of denaturation at 94°C for 30 sec, annealing 55–62°C depending on the primer pair requirements for 30 sec, extension at 72°C for 30 sec, and a final extension at 72°C for 3 min. The PCR was carried out in a GeneAmp 9600 PCR system (ABI), and PCR products were analyzed on 2% 1× TAE agarose gels. DNA for CEPH mega-YACs was isolated as reported (Tsaur et al. 1995). Assays for STS content of mega-YAC clones were conducted in 25-ml PCR reactions as above.
Acknowledgments
We thank Markus Stoffel for useful discussion, Lisa Winick for proofreading the manuscript, and Susan Korres for assistance in preparing the manuscript. These studies were supported by funds from the Howard Hughes Medical Institute and a grant from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵1 Corresponding author.
-
E-MAIL friedj{at}rockvax.rockefeller.edu; FAX (212) 327-7420.
-
- Received February 24, 1998.
- Accepted July 28, 1998.
- Cold Spring Harbor Laboratory Press