New Insulin-Like Proteins with Atypical Disulfide Bond Pattern Characterized in Caenorhabditis elegans by Comparative Sequence Analysis and Homology Modeling
- 1Laboratoire BGBP–UMR Centre National de la Recherche Scientifique (CNRS) 5558, Université Claude Bernard - Lyon 1, F-69622 Villeurbanne Cedex, France; 2Geneva Biomedical Research Institute, Glaxo-Wellcome, CH-1228 Plan-les-Ouates, Switzerland; 3Département de Biochimie Médicale, CH-1211 Geneva 4, Switzerland
Abstract
We have identified three new families of insulin homologs inCaenorhabditis elegans. In two of these families, concerted mutations suggest that an additional disulfide bond links B and A domains, and that the A-domain internal disulfide bond is substituted by a hydrophobic interaction. Homology modeling remarkably confirms these predictions and shows that despite this atypical disulfide bond pattern and the absence of C-like peptide, all these proteins may adopt the same fold as the insulin. Interestingly, whereas we identified 10 insulin-like peptides, only one insulin-like-receptor (daf-2) has been found. We propose that these insulin-related peptides may correspond to different activators or inhibitors of the daf-2insulin-regulating pathway.
Insulin and related peptides are key hormones for the regulation of growth and metabolism. Originally discovered in mammals, insulin-related peptides have been identified in chordates, mollusks, and insects. Recently, an insulin receptor-like gene,daf-2, has been reported in the nematode Caenorhabditis elegans (Kimura et al. 1997). These investigators showed that as in mammals, this gene is involved in the regulation of metabolism. Interestingly, they also showed that this gene is necessary for the nematode to enter into diapause and that it affects its longevity. Thus, their finding not only demonstrate that the genetic circuitry that regulates glucose metabolism was already present in the last common ancestor of mammals and nematodes, >600 million years ago, but also suggests a possible link between aging and glucose metabolism. Two downstream components of this insulin-like signaling pathway have been already described: the kinase age-1 (Morris et al. 1996) and the transcription factor daf-16 (Ogg et al. 1997). However, the ligand(s) that activate this pathway are still unknown. To identify potential daf-2 ligands, we analyzed protein sequences issued from C. elegans genome project (Wilson et al. 1994) to search for insulin-related peptides. Here we report the discovery of 10 insulin homologs in C. elegans that are potential ligands for daf-2. We have identified several concerted mutations indicating that despite overall low sequence similarity and an atypical disulfide bond pattern, these proteins adopt the same tertiary structure as other insulin-related peptides. This finding is confirmed by structure prediction of these proteins by comparative modeling.
RESULTS
Search for Insulin-Related Proteins
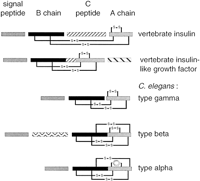
Insulin-related proteins are grouped together on the basis of strong structural similarity. They are all characterized by a signal peptide, a B-chain, a connecting C-peptide, and an A-chain. Chains B and A contain, respectively, two and four strictly conserved cysteines, involved in three disulfide bonds: two between chains A and B, and a third within the A-chain (Fig. 1). The C-peptide connecting B and A chains in the propeptide is removed by proteolytic cleavage, except in insulin-like growth factors (IGF I and IGF II) in which the C-domain is retained in the mature peptide. Vertebrate IGFs and amphioxus insulin-like peptide (ILP) are also characterized by an additional carboxy-terminal domain.
Schematic representation of vertebrate insulin, IGF, and C. elegans insulin-like predicted proteins. Domains that are cleaved during maturation of the propeptide are separated by spaces. Disulfide bonds are indicated. C. elegans insulin-like proteins are characterized by the absence of C-peptide. Type β and type α have no potential cleavage site between B and A chains; type β and type α contain an additional disulfide bond between B and A chains. In type-α proteins, the disulfide bond within the A chain is substituted by hydrophobic interaction between Phe and/or Tyr residues.
Currently, seven families can be recognized among insulin-related peptides: (1) insulin, IGF I and IGF II from vertebrates, and amphioxus ILP (Chan et al. 1990); (2) bombyxin brain secretory peptides fromBombyx mori (insect) (Kondo et al. 1996); (3) molluscan insulin-related peptides 1–7 (MIP) (Smit et al. 1996); (4) vertebrate relaxins (Bullesbach et al. 1986); (5) mammalian Leydig cell-specific insulin-like peptide (LeyI-l, INSL3) (Burkhardt et al. 1994); (6) insulin-related peptide (LIRP) from locust (insect) (Lagueux et al. 1990); and (7) mammalian early placenta insulin-like peptide (EPIL, INSL4) (Chassin et al. 1995).
The search for new insulin-related peptides is made difficult by the fact that their primary sequence is poorly conserved. Hence, we used the profile technique, a sensitive approach for sequence database searches (Bork and Gibson 1996). Two profiles, corresponding to the conserved B and A chains, were constructed from an alignment of representatives of the different currently identified insulin-like families. Two iteration cycles of profile searches in SWISS-PROT and TREMBL [cumulative updates, August 9, 1997 (Bairoch and Apweiler 1997)] identified nine new proteins, all from C. elegans,that match both to B and A chains, in the proper orientation and with a high statistical significance (P < 2 × 10−5). We found several other potential homologs, but matching only the A chain and with a lower statistical significance. These borderline cases will not be discussed here.
These nine proteins are currently described in databases as hypothetical proteins, predicted from computer analysis of genomic sequences. One of these hypothetical proteins (ZK84.3, SWISS-PROT accession no. Q23631) contained in its amino-terminal part a duplication of the region spanning the signal peptide and the B domain. In the genomic sequence (EMBL accession no. U23181) these duplicated regions correspond to two exons, separated by a 1.7-kb intron, which is exceptionally long for C. elegans (Blumenthal and Spieth 1996). Sequence analysis revealed that this predicted intron contained a putative exon, 53 nucleotides downstream of the first exon, in the same phase, and encoding an A-like carboxy-terminal domain. Therefore, we think that the predicted ZK84.3 gene is an artifact caused by an error in exon prediction and that there are two genes in tandem, both coding for complete insulin-related peptides [this correction has been approved by the authors of the genomic sequence (B. Waterson, pers. comm.)]. These two predicted genes will hereafter be referred to asZK84.3_1 ( P56173 ) andZK84.3_2 ( P56174 ). The 10 C. elegans insulin-related peptides that we have identified are indicated in Figure 2.
Alignment of C. elegans ILPs with representatives of all currently identified insulin-related families. (INSL4) Human early placenta ILP (EPIL); (INSL3) human Leydig ILP (LEY I L); (RLN1) human prorelaxin H1; (BBXA, BBXB, BBXC, BBXD) B. mori bombyxin (BBX) A9, B1, C2, and D1; (LIRP) locust insulin-related peptide. (MIP I, MIP II, MIP III, MIP V, MIP VII) Molluscan insulin-related peptides; (ILP) Amphioxus ILP; (INS) human insulin; (IGF1, IGF2) human insulin-like growth factor I and II (IGF II). C. elegans insulin-like peptides: (C17C3.4) γ type; (ZK1251.2, ZK75.1, ZK75.2, ZK75.3, ZK84.3_1, ZK84.3_2: β-type; (M04D8.1, M04D8.2, M04D8.3) α type. (*) Positions of cysteines involved in the additional disulfide bond between B and A chains of α and β types. Daggers indicate positions of Phe or Tyr substituting the disulfide bond within A chain of α type; (C) conserved cysteines; (•) other conserved positions. SWISS-PROT/TREMBL accession nos. are shown. The length of the nonconserved peptide linking B and A chains is indicated.
C. elegans Insulin-Related Proteins Evolved by Gene Duplication
Besides their similarity in B and A domains, these 10 proteins share several features with the other insulin-related peptides: In all cases the propeptide is relatively short (76–112 amino acids) and is predicted to contain a signal peptide [by use of SignalP program (Nielsen et al. 1997)], as expected for secreted peptides. Interestingly, all of the genes encoding these proteins contain one intron, between the exons coding for the B and A domains, in phase 1 (i.e., splitting the reading frame between positions 1 and 2 of a codon). Thus, these genes have exactly the same structure and intron phase as all the other currently identified insulin-related genes, except for the bombyxin genes that have no intron (Kondo et al. 1996). This latter argument clearly shows that all these genes are true homologs, that is, the similarity between the encoded proteins is not caused by convergent evolution but by common ancestry.
These 10 C. elegans genes are located in four different loci: a cluster of five contiguous genes in chromosome II (ZK75.2, ZK75.3, ZK75.1, ZK84.3_1, ZK84.3_2), a cluster of three contiguous genes in chromosome III (M04D8.1, M04D8.2, M04D8.3),ZK1251.2 in chromosome IV, and C17C3.4 in another locus of chromosome II. By phylogenetic analysis, we identified three subfamilies among these proteins: one corresponding to chromosome III cluster (type α), another to chromosome II cluster and toZK1251.2 (type β), and a third represented byC17C3.4 (type γ) (Fig. 2). Proteins from the three different subfamilies are highly divergent among each other (13%–25% identity in A and B chains), suggesting two ancient duplication events, followed by successive tandem duplications within both clusters of chromosome II and III. (For comparison, vertebrate insulin and amphioxus ILP that diverged ∼500 million years ago share 50% identity in A and B chains.)
Structure of C. elegans Insulin-Related Proteins
Four noteworthy observations can be made as to the structure of these putative insulin-like proteins:
- 1.
- The six proteins of the type-β subfamily (ZK75.2, ZK75.3, ZK75.1, ZK84.3_1, ZK84.3_2, and ZK1251) contain a short additional domain (29–39 amino acids), between the signal peptide and the B domain (Fig. 1). This extra domain ends up with a basic dipeptide and is probably cleaved from the mature peptide.
- 2.
- In all 10 proteins the equivalent of a C propeptide that is found in all characterized members of this family between the B and the A chains is absent. In type-α and -β families, there is no basic dipeptide between the B and A domains. Thus, it is tempting to propose that type-α and -β proteins are probably not processed in two separate chains but rather form a single polypeptide (Fig. 1).
- 3.
- The four cysteines involved in disulfide bonds between A and B domains are conserved in all C. elegans insulin-like proteins. Interestingly, proteins of type α and β have two additional cysteines, one located at the carboxy-terminal extremity of the B chain-like domain and the second at the carboxyl terminus of the A chain-like domain (Fig. 2). These correlated occurrences suggest that these cysteines could participate in an extra disulfide bond.
- 4.
- Like all currently known insulin-like peptides, type-β and -γ families contain two cysteines linked by a disulfide bond within the A domain. These two cysteines are absent in type-α insulin-like proteins, in which they are substituted by phenylalanine (F) and/or tyrosine (Y) residues. Three different combinations are observed: F/Y in M04D8.1, Y/F in M04D8.2, and Y/Y in M04D8.3 (Fig. 2). These concerted mutations suggest a direct interaction between these two residues, which might compensate for the absence of disulfide bond within the A domain.
To test these three latter hypotheses (2–4), the tertiary structures of the mature peptides ZK75.3 (type β, Q09628, residues 56–108), M04D8.3 (type α, Q21506, residues 19–76) and M04D8.2 (type α, Q21508, residues 27–83) were predicted by comparative modeling from the human IGF1 (PDB entry 3gf1) (Cooke et al. 1991) by use of a combination of Swiss-PdbViewer (Guex and Peitsch 1996), and SWISS-MODEL (Peitsch et al. 1995). CHARMm (Brooks et al. 1983) was used to optimize the stereochemistry of the models, as described previously (Peitsch 1996).
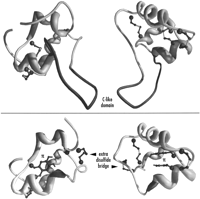
In IGF-1, the last residue of the B domain and the first residue of the A domain are spatially close (CA–CA distance of 3.7 Å between Pro-27 and Gly-42 in the first NMR model contained in the PDB entry 3gf1). Thus, removing the C-like domain by making a direct bridging of domains B and A could easily be obtained in the predicted structures (Fig. 3).
Comparison of human IGF-1 (top; PDB entry 3gf1, NMR model 4) with a model of type-α M04D8.3 ILP of C. elegans(bottom; SWISS-PROT entry Q21506). Noteworthy differences between the two proteins are mentioned: The C-like domain present in IGF-1 only (darker), the supplementary disulfide bridge present inC. elegans (arrowhead), and the replacement of one of the IGF-1 disulfide bridges by an interaction of two Tyr in C. elegans (π). Two different views are presented for a better understanding of the differences.
Quite remarkably, the CA–CA distance between the residues corresponding to the two extra cysteines characteristic of the α and β types is of 6 Å, which is the optimal distance for a disulfide bridge. As a matter of fact, an additional disulfide bridge between B and A domains was readily obtained simply by mutating Gly-22 and Ala-62 of IGF-1 to Cys within Swiss-PdbViewer (Guex and Peitsch 1996).
As mentioned earlier, in type α, the two cysteines involved in the intrachain disulfide bridge in IGF-1 are substituted by the much larger aromatic amino acids Tyr or Phe. Those could be accommodated with a slight displacement of the backbone (1 Å). These two aromatic residues can be positioned to make a π stacking that could partially compensate for the loss of the disulfide bridge (Fig. 3). This configuration was favored, as it allows an optimal occupancy of space, creating a strong hydrophobic core in the structure.
The models have been deposited in the SWISS-MODEL repository (http://www.expasy.ch/swissmod/accession nos. Q21506, Q21508, and Q09628).
It should be noted that the protein encoded by ZK84.3_1, which clearly belongs to the β-type family, does not contain the first of the two cysteines involved in the additional disulfide bond between B and A domains. However, several other features suggest that this gene might be nonfunctional: (1) There is a deletion of one residue in the B domain (Fig. 2); (2) several highly constrained residues of the B domain are not conserved (Fig. 2); (3) the amino-terminal domain does not seem to be cleavable; and (4) the intron splice site does not fit the GT–AG consensus. Thus, it is likely that this is a pseudogene or that it encodes a peptide that has lost its original function.
DISCUSSION
We have identified three new families of insulin-like peptides (α, β, and γ types) represented by 10 proteins in the nematode C. elegans. These proteins are the result of ancient gene duplications and are clearly homologous to other insulin-like proteins. However, they are atypical in that they all lack a C-like peptide. Moreover, we identified several concerted mutations that suggest that in α and β types, an additional disulfide bond links B and A-like domains, and that in α type, the disulfide bond within the A-like domain is substituted by a hydrophobic interaction between tyrosines and/or phenylalanines (Figs. 1 and 3). Comparative modeling shows that despite these atypical features, the C. elegans proteins can adopt the same tertiary structure as the other insulin-like peptides. Quite remarkably, the residues for which we noticed correlated mutations are very close in the predicted structures. Thus, we believe that the interactions that we predicted really occur in vivo.
In α- and β-type insulin-like peptides, the absence of basic dipeptide between B and A domains suggests that the peptide is not cleaved in two chains. Such is also the case for vertebrate IGFs, but the IGFs have conserved a fossil C-type region. The absence of sequence conservation between the C peptides in the different insulin subfamilies has always been said to be caused by a low evolutionary pressure on the sequence itself, but its presence was important for the proper folding and disulfide topology of the mature protein. However, our models show that in spite of the absence of a C-like domain, theC. elegans peptides can adopt the same fold as the other insulin-related peptides. It is interesting to note that the additional disulfide bridge between B and A domains that we have found in α and β types is located in the same region of the protein as the C-like loop (Fig 3). Thus, this disulfide bridge might be required for the protein to fold properly in the absence of a C-like domain.
Our finding of insulin homologs in C. elegans shows that a primordial insulin-type gene existed in the chordate/nematode ancestor, thus confirming that the insulin signaling pathway was already present very early in metazoan evolution (see note). However, we did not detect any insulin-related protein in plants or in fungi, even though the complete yeast genome and a large amount of plant data are available. This suggests that this signaling pathway is probably specific to metazoans.
It is interesting to note that, whereas we have found three families of insulin-like peptides (represented by 10 different proteins, among which 1 is probably nonfunctional), only one insulin-receptor related gene has been identified in the C. elegans genome (Kimura et al. 1997). To date, ∼70% of C. elegans genome (100 Mb) has been sequenced (http://www.sanger.ac.uk/Projects/C_elegans/August1997). Thus, it is possible that other insulin-like receptors remain to be discovered in C. elegans. Another possible explanation is that these different insulin-like peptides might compete to bind the unique receptor daf-2, some of them acting as activators, others acting as inhibitors. It is also important to note that this apparent redundancy may explain why, unlike daf-2, age-1, anddaf-16 (Morris et al. 1996; Kimura et al. 1997; Ogg et al. 1997), these insulin-related genes have not been identified by classical genetic approaches.
Despite evolutionary distance, C. elegans is a good model organism. Thus, the finding of insulin homologs, potential ligands fordaf-2, should be very valuable to understand the insulin-regulating pathway, not only in C. elegans, but also in mammals.
METHODS
Profile searches were performed with the PFSEARCH program (P. Bucher 1997, pftools release 2.0, available fromftp://ulrec3.unil.ch/pub/pftools), which implements the method described by Bucher et al. (1996). The statistical significance was estimated following the procedure described by Hofmann and Bucher (1995).
Acknowledgments
This work is supported by the CNRS.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
NOTE
An insulin gene from the sponge Geodia cydonium has been reported previously (Robitzki et al. 1989). However, we have shown by phylogenetic analysis that the DNA sequence coding for this protein does not come from sponge but probably from a contamination by rodent material, as confirmed by the investigators (W.E.G. Muller, pers. comm.).
Footnotes
-
↵4 Corresponding author.
-
E-MAIL duret{at}biomserv.univ-lyon1.fr; FAX 33 4 78 89 27 19.
-
- Received November 18, 1997.
- Accepted February 17, 1998.
- Cold Spring Harbor Laboratory Press