Mapping the Germ-Line and Somatic Genomes of a Ciliated Protozoan,Tetrahymena thermophila
Abstract
Ciliates are among the very few eukaryotes in which the powers of molecular biology, conventional genetics, and microbial methodology can be synergistically combined. Because ciliates also are distant relatives of vertebrates, fungi, and plants, the sequencing of a ciliate genome will be of import to our understanding of eukaryotic biology. Tetrahymena thermophila is the only ciliate in which a systematic genetic mapping of DNA polymorphisms has begun.Tetrahymena has many biological features that make it a specially or uniquely useful experimental system for fundamental research in cell and molecular biology and for biotechnological applications. A key factor in the usefulness of Tetrahymena is the speed, facility, and versatility with which it can be cultivated under a wide range of nutrient conditions, temperature, and scale. This article describes the progress made in genetically and physically mapping the genomes of T. thermophila: the micronuclear (germ-line) genome, which is not transcriptionally expressed, and the macronuclear (somatic) fragmented genome, which is actively expressed and determines the cell’s phenotype.
Introduction to the Ciliated Protozoa
Tetrahymena is a member of the Ciliated Protozoa, a monophyletic group of unicellular eukaryotes, whose biology was reviewed recently in Hausmann and Bradbury (1996). The ciliates diverged earlier than plants and fungi in the evolutionary line leading to the vertebrates (see Pace 1997). Yet the ciliates possess a typical eukaryote life cycle, with conventional meiosis and biparental fertilization through the union of haploid gametes. While retaining unicellularity, ciliates exhibit evolutionary advances reminiscent of features of multicellular eukaryotes. For example, ciliates possess vegetative growth restricted to the diploid phase of their life cycle; extensive compounding of cellular structure that has led to the evolution of macroscopically observable unicells; elaborate structural differentiations accompanied by complex morphogenetic mechanisms; internal fertilization through direct exchange of gamete nuclei; and nuclear dimorphism, that is, the possession of differentiated germ-line and somatic nuclei, described below.
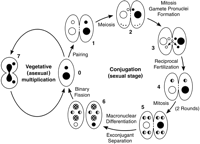
The possession of two related but functionally differentiated genomes within one cell is a diagnostic ciliate feature. The germ-line genome is carried in the diploid micronucleus (MIC), whereas the somatic genome is contained in the polyploid macronucleus (MAC). MIC and MAC differentiate from mitotic descendants of the fertilization nucleus formed during conjugation, the sexual stage of the life cycle (Fig.1). MAC differentiation is accompanied by chromosome fragmentation and thousands of other DNA rearrangements (Prescott 1994;Coyne et al. 1996). These rearrangements, which are developmentally programmed and site-specific, reconfigure the genome extensively. Nuclear differentiation results in a MIC that is not transcriptionally expressed and an actively expressed MAC that determines the cell’s phenotype. The nuclear dualism of the ciliates is accompanied by a remarkable set of cellular and molecular mechanisms, and generates unusual experimental genetics capabilities as well.
Generalized ciliate life cycle. (0) Vegetative cells. (Small and large circles) MIC and MAC, respectively. (1) Two paired cells, homozygous for alternative alleles at one locus. (2) MICs undergo meiosis, and four haploid nuclei are produced. Only the anterior meiotic product remains functional; the other three disintegrate. This is the stage at which meiotic crossing-over, used for genetically mapping the MIC genome, occurs. (3) Mitotic division of functional meiotic product yields genetically identical migratory (anterior) and stationary (posterior) gamete pronuclei. (4) Migratory pronuclei are reciprocally exchanged and fuse with stationary pronuclei of the recipient cell, forming the zygote nucleus, which is diploid and, in this instance, heterozygous. (5) The zygote nucleus undergoes two mitotic divisions, giving rise to four genetically identical diploid nuclei. (6) Two of those nuclei (checkerboard-filled) have differentiated into macronuclei; the other two (solid and white halves) remain diploid micronuclei. The old MACs (at the bottom of each conjugant) are being resorbed and will be lost. This is the stage at which chromosome fragmentation and other site-specific DNA rearrangements occur in the differentiating MAC. The two exconjugants have separated and undergo their first binary fission, restoring the normal nuclear composition (back to stage 0). (7) Vegetative cell dividing by binary fission. The diploid MIC has divided mitotically; the polyploid MAC is undergoing “amitotic division,” pinching off into roughly equal halves. This life cycle scheme is highly conserved among ciliates, although differences of detail occur in particular groups and species.
Research in ciliate molecular, genetic, and developmental cell biology has focused predominantly on members of two evolutionary groups that branched out early in the evolution of the ciliates:Paramecium and Tetrahymena in one branch, andEuplotes, Oxytricha, and Stylonychia in another. Modern genetics of unicellular eukaryotes began withParamecium after Sonneborn (1937) discovered mating types. The contributions made by Paramecium genetics have been reviewed by Preer (1997). Although Mendelian genetic processes have been demonstrated in a variety of ciliate genera, conventional genetic studies have been done mostly in Tetrahymena (founded byNanney and Caughey 1953; for review, see Orias 1998, with an extended version in Orias 1997) and in Paramecium. A wide variety of useful laboratory mutants have been obtained in both genera. DNA-mediated transformation has been accomplished in Paramecium (see below), Tetrahymena (Gaertig and Gorovsky 1995; Cassidy-Hanley et al. 1997), and Stylonychia (Ascenzioni and Lipps 1986). Gene replacements and knockouts have been done in theTetrahymena MIC and MAC (e.g., Hai and Gorovsky 1997). Gene cloning by complementation has been accomplished in Paramecium(e.g., Skouri and Cohen 1997).
Tetrahymena Genome Initiative
Tetrahymena has many special features that make it a specially or uniquely useful experimental system for fundamental research in cell and molecular biology and for biotechnological applications (Box ; see also Nanney 1980). In addition to being amenable to a wide repertoire of genetic and molecular manipulation, a key factor in the usefulness of Tetrahymena is the speed, facility, and versatility with which it can be cultivated. At <2 hr, its doubling time ranks with that of the fastest eukaryotic microbes. It grows efficiently using bacteriological methods under a wide variety of conditions: in axenic (i.e., pure culture) conditions, either in rich media or in nutritionally complete, totally defined synthetic medium (Wheatley et al. 1994), or in bacteria-containing media. It grows in volumes ranging from microdrops to industrial fermentors (Kiy and Tiedtke 1992), throughout a widetemperature range—from 12°C to 41°C.
Selected contributions to fundamental research and potential biotechnological applications exploiting unique or special features of the biology of Tetrahymena and other ciliates1. Current research on general problems of fundamental significance. • Ribozymes: Codiscovery in the self-splicing intron of the 28S ribosomal RNA (Kruger et al. 1982). TheTetrahymena macronucleus as the eukaryotic nucleus most actively synthesizing RNAs. • Eukaryotic telomeres: Discovery of telomere molecular basis (Blackburn and Gall 1978) and telomerase (Greider and Blackburn 1985); template role of telomerase RNA (Yu et al. 1990) in Tetrahymena (see alsoBlackburn and Greider 1995). Tetrahymena has 20,000 copies of the DNA telomere per cell and, during MAC differentiation, has high levels of telomerase activity to carry out rapid de novo synthesis of >4000 telomeres. • Chromatin structure and function (e.g., Roth and Allis 1996; Shen and Gorovsky 1996): The ciliates represent an experiment of nature, as they maintain within the same cell a MIC, with chromatin always condensed and not expressed, and a MAC, with chromatin always extended and actively expressed. • Site-specific chromosome diminution: Mechanisms of diminution as well as understanding of epigenetic control and transposon relationships in Tetrahymenaand other ciliates (e.g., Prescott 1994; Coyne et al. 1996; Meyer and Duharcourt 1996; Klobutcher and Herrick 1997). Thousands of site-specific diminution events occur per haploid genome during MAC differentiation. • Regulated secretion (e.g.,Chilcoat et al. 1996 in Tetrahymena; Madeddu et al. 1994 inParamecium): These ciliates have thousands of secretory vesicles; their release can be induced in large number and synchronously. • Membrane excitability and chemosensory signaling in Paramecium (e.g., Saimi et al. 1994; Van Houten 1994): These large cells have striking and easily observed swimming behavior that indicates the state of depolarization of the membrane. • Cytokines: Studies are facilitated inTetrahymena by availability of nutritionally complete, chemically defined growth media (e.g., Rasmussen et al. 1996); inEuplotes (e.g., Luporini et al. 1996), where the easily observable mating reaction is induced by pheromones secreted in large amounts. • Cellular morphogenesis and polarity control (e.g., Frankel 1989): In addition to available genetic approaches, ciliates can be surgically manipulated. • Role of post-translational modifications of tubulin in Tetrahymena (e.g., Gaertig et al. 1995): Tetrahymena possesses a remarkable variety of tubulin-based structures formed from a minimal number of tubulin genes. 2. Applications related to nutritional, environmental, and occupational health sciences based onTetrahymena as a eukaryote useful for rapid, reliable, sensitive, and inexpensive bioassays. • Determining the protein nutritional value of human foods, based on similarity of nutritional requirements (e.g., Koehler et al. 1987). • Sensitive biosensor for biotoxin detection (e.g., Martin Gonzalez et al. 1997). • Determining quantitative structure-activity relationship (QSAR) for the toxicity of human-made chemical compounds in the environment or the pharmacopoeia (e.g., Pauli et al. 1994; Sinks et al. 1997). • Monitoring water quality (e.g., Slabbert and Morgan 1982). • Alternative host for the growth ofLegionella, the causative agent of Legionnaire’s disease (e.g., Kikuhara et al. 1994). • Alternative to vertebrate animals or cells for testing cleaning products for eye irritation (e.g., Bagley et al. 1994). 3. Potential for biotechnological applications. • Industrial production of enzymes and pharmaceuticals (e.g., Ropenga and Lenfant 1987; Kiy et al. 1996). • Biological control of mosquitoes, intermediate hosts for major worldwide human diseases: malaria, yellow fever (e.g., Manasherob et al. 1996; Narain et al. 1996). • Fighting epidemics of the closely related ciliate Icthyophthirius in fish hatcheries and pet stores (e.g., Ling et al. 1993).
Tetrahymena thermophila is the only ciliate in which a systematic genetic mapping of DNA polymorphisms has begun (Orias 1997). An important short-term aim of the genome mapping initiative has been to facilitate the cloning of biologically interesting mutantTetrahymena genes when molecular probes are not available and cloning becomes dependent on genetic map location. The long-term aim has been to build a genetic framework that will anchor physical maps of the two Tetrahymena genomes and eventually the genome sequence.
Mapping the Tetrahymena MIC Genome by Meiotic Recombination
Physical Basis of the MIC Genome
The germ-line (MIC) genome of Tetrahymena has a complexity of ∼220 Mb of DNA (Karrer et al. 1986), one order of magnitude lower than that of the human genome. The MIC contains five pairs of metacentric chromosomes of roughly equal size, readily visible at the metaphase stages of meiosis and postzygotic mitoses during conjugation. Although there are some repetitive sequences, most of the DNA is present in single copy.
Rationale for Constructing the MIC Genetic Map
Tetrahymena genome mapping has so far been mainly based on DNA polymorphisms obtained by the randomly amplified polymorphic DNA (RAPD) method described by Williams et al. (1990). Two inbred strains of T. thermophila, B and C3, have been used as the source of polymorphisms. The maps of MIC linkage groups have been constructed on the basis of meiotic recombination frequency, as has been done for nearly a century for eukaryotes possessing meiosis in their life cycle (Griffiths et al. 1996). The application of mapping rationale and RAPD methodology to Tetrahymena has been described (Lynch et al. 1995; Brickner et al. 1996). Special features of Tetrahymenagenetics made possible by nuclear dualism have facilitated the mapping: (1) viability of strains missing both copies of particular chromosomes in the MIC; (2) viability of aneuploid monosomic strains, made possible by copy number regulation in the polyploid, fragmented MAC; and (3) ability to obtain meiotic segregants that are homozygous for their entire genomes (MIC and MAC) in a single step (Brickner et al. 1996).
Current Status of the Tetrahymena Genetic Map MIC
Nearly 400 RAPDs have been assigned to linkage groups with high statistical confidence, that is, with log of the odds (lod) against independent segregation >3. These RAPDs fall into 26 linkage groups and are distributed among all the chromosome arms (Fig.2). The use of monosomic strains has allowed—and has provided a running check on—the assignment of linkage groups to chromosome arms. Of the RAPDs tested, 94% are linked to at least one other RAPD. A total of 4450 cM have been linked (Table1). Given that the MIC genome is 220 Mb and that more centiMorgans probably remain to be linked, the average rate of meiotic recombination is at most 50 kb/cM. This value already represents a much higher average rate of meiotic recombination than that in most eukaryotes, for example, 20 times higher than the average rate for the human genome.
Maps of the MIC genome, by chromosome arm, based on ∼400 RAPD polymorphisms. (L and R) Left and right chromosome arms, except for chromosome 5, where arms cannot yet be distinguished. (U) Linkage group with uncertain chromosome assignment. Chromosomes throughout this review are numbered according to the recently revised scheme (J. Merriam, D. Cassidy-Hanley, and P.J. Bruns, pers. comm.). (•) The centromere region of chromosome 4, as RAPDs on its left and right map to 4L and 4R, respectively, by monosomic mapping. Other linkage groups within a chromosome arm remain to be oriented with respect to one another or to the centromere; this will be accomplished when more DNA polymorphisms are mapped or when immunocytological mapping inTetrahymena (D. Cassidy-Hanley and P.J. Bruns, pers. comm.) is used with the existing RAPDs. Most RAPDs have been mapped using only 32 meiotic segregants. Within a given linkage group, the order within a locus cluster and the orientation of clusters separated by large distances are still soft: Inversions often change the likelihood of the segregation data by <3 log units. Clusters of loci that failed to give recombinants with one another are represented by a single point. Loci unlinked to all others are not shown.
Summary of centiMorgans Linked by MIC Chromosome Arms
We have no rigorous way to estimate what fraction of the genome has already been mapped. However, if we take the average number of centiMorgans in chromosomes 1 and 4—that is, those with the highest linked centiMorgans (Table 1)—and multiply it by 5, we arrive at an estimated 5680 cM for the whole genome. This suggests that up to 78% of the genome may have been genetically linked so far.
Mapping Classical Loci and Cloned Genes by Meiotic Recombination
Many Tetrahymena loci classically described on the basis of biological phenotypes, cloned DNA genes, and MIC-limited DNA segments have been mapped to chromosome arms (Bruns and Cassidy-Hanley 1993; Cassidy-Hanley et al. 1994). Some of the classical loci have been assigned to linkage groups (Allen et al. 1996; Brickner et al. 1996; C.B. Van Slyke, E. Orias, and E.P. Hamilton, unpubl.). GenBank files contain sequences for >50 T. thermophila cloned genes. A collaborative effort is being coordinated by the Orias laboratory to map cloned Tetrahymena genes to linkage groups. The approach consists of identifying a B-C3 RFLP associated with each gene and then mapping the RFLP using the standard segregant panels and databases.
Mapping the Tetrahymena MAC Genome
Physical Basis of the Tetrahymena MAC Genome
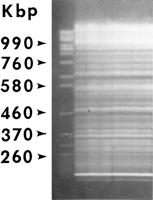
The MAC genome is a fragmented, diminished, and greatly amplified version of the MIC genome. The site-specific DNA rearrangements that generate the MAC genome occur only once per life cycle, that is, when new MACs are differentiated from mitotic sisters of the MICs during the second half of conjugation. The MAC genome consists of ∼270 autonomously replicating pieces (ARPs). These ARPs (Fig.3) range between 21 kb (the ARP that carries the rRNA genes) and >1 Mb, with an average estimated at ∼800 kb (Altschuler and Yao 1985; Conover and Brunk 1986). Most, if not all, of these DNA ARPs are generated by site-specific fragmentation of the germ-line-derived genome at chromosome breakage sites (Cbs). The Cbs, a 15-mer of double-stranded DNA, is necessary and sufficient for fragmentation (Yao et al. 1990). The Cbs system in effect generates in the MAC a self-reproducing natural equivalent of a rare-cutter restriction digest of the MIC genome. Telomeres are added de novo at the ends of newly generated DNA ARPs by telomerase. Each of these ∼270 ARPs are amplified at the time of MAC differentiation to 45 copies per MAC, with the exception of the rDNA ARP, which is turned into an inverted (palindromic) repeat and is amplified to ∼10,000 copies per MAC (Kapler 1993). ARPs lack kinetochores or any other means of regular distribution. Their copies (and the gene alleles they carry) are distributed at random during MAC division.
The fragmented MAC genome of T. thermophila, visualized by pulse-field gel electrophoresis of whole-cell DNA. (Left lane) The chromosomes of Saccharomyces cerevisiae, used as size markers. Brighter bands most likely represent coincidences of unrelated ARPs of the same size, except for the two lowest bands—the mitochondrial and MAC rDNA. MIC DNA stays in the wells under the conditions used.
An estimated 10%–20% of the germ-line-derived genome is lost during differentiation of the Tetrahymena MAC (see Karrer 1986). Some loss occurs in conjunction with chromosome fragmentation. Most of the loss occurs through site-specific deletion of internal segments (Yao 1996), of which there are an estimated 6000 per haploid MIC genome. Germ-line DNA segments can thus be classified as MIC-limited or MAC-destined, depending on their fate at MAC differentiation. Internal segment elimination in Tetrahymena is accompanied by the preferential loss of repeated DNA.
Rationale for Mapping the MAC Genome by Assortment
Tetrahymena is the first organism in which genetic mapping of a fragmented somatic eukaryotic genome has become possible, because of the random distribution of ARP copies and the relative rarity of crossing-over in the MAC. In initially heterozygous MACs, the random distribution of allele copies generates, during subsequent MAC divisions, a genetic drift that results in MACs pure for one or the other allele. This phenomenon is known as macronuclear assortment and was discovered and analyzed by Allen and Nanney (1958). Heterozygotes propagated vegetatively for 300–500 fissions have high probability of having completed assortment at any given locus (Doerder et al. 1975) and have been dubbed “terminal assortants” (Longcor et al. 1996).
Terminal assortants of cells initially heterozygous at two loci almost always exhibit one parental genotype when the loci are carried on the same MAC ARP (Longcor et al. 1996), that is, they exhibit nearly complete coassortment. This result defines a coassortment group. In contrast, two loci on different ARPs assort independently, that is, there is no statistical excess of parental types among terminal assortants. Rigorous molecular genetic evidence has been provided that ARPs are the physical basis of coassortment groups (L. Wong, L. Klionsky, N. Nangle, D. Mendinueto, V. Merriam, S. Wickert, J.D. Orias, E. Orias, and E. Hamilton, in prep.). Coassortment is thus the MAC analog of MIC linkage and provides a basis for determining whether or not two polymorphic loci are carried in the same MAC ARP. The mapping of the macronucleus to physically defined entities by purely genetic means represents an important recent advance in Tetrahymena genetics.
RAPDs eliminated from the MAC, either in connection with chromosome fragmentation or as internally eliminated segments, can also be detected genetically (see Longcor et al. 1996). Because these RAPDs reside exclusively in the MIC, with its regular (mitotic) distribution of alternative allele copies, such RAPDs do not show the assortment typical of those in MAC-destined segments.
Current Status of MAC Genome Mapping
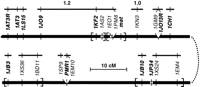
A clear distinction between loci that assort independently and loci that show a strong tendency to coassort has been found so far (Longcor et al. 1996; L. Wong , L. Klionsky, N. Nangle, D. Mendinueto, V. Merriam, S. Wickert, J.D. Orias, E. Orias, and E. Hamilton, in prep.). To date, 32 coassortment groups that contain at least two loci have been identified, originating from all the MIC chromosomes. MIC chromosome 1 is the one mapped most extensively in the MAC; a segment of the current coassortment map of its left arm is shown in Figure4. The ratio of coassortment groups to mapped loci, if representative, suggests that the RAPDs so far mapped to the MIC will fall into ∼170 coassortment groups, which will represents 63% of the estimated total of 270 ARPs (L. Wong, L. Klionsky, N. Nangle, D. Mendinueto, V. Merriam, S. Wickert, J.D. Orias, E. Orias, and E. Hamilton, in prep.). In most if not all cases, MIC and MAC maps are colinear, that is, coassorting loci map to continuous segments in the MIC map. This is in agreement with the notion that ARPs are generated simply through Cbs-determined fragmentation. Coassortment mapping thus provides a totally independent check on the close linkage of loci in the MIC. Rare exceptions to colinearity, in which ARPs are generated by joining together discontinuous MIC segments, may exist. In at least one such potential case, increasing local map resolution by testing more meiotic segregants has led to a reordering of the MIC map and has restored colinearity.
Coassortment map of a segment of the left arm of chromosome 1. (Thick bar) A continuous MIC chromosome segment; (open segments) MIC-limited sequences, absent from the MAC; (tick marks) genetic loci. The order of loci shown in boldface type has been determined with log odds >3.0; the order of loci within brackets remains uncertain at that level of statistical significance. Loci emanating from a single point so far have shown no recombination with one another. (Solid thin bars) MAC coassortment groups, such that two loci within a group coassort with one another, and two loci on different groups assort independently of one another. The sizes of two coassortment groups already physically related to ARPs (Longcor et al. 1996) are shown in megabase pairs. Technical description of the DNA polymorphisms is available in Orias (1997).
Physical mapping of coassortment groups to MAC ARPs, to determine their size and restriction pattern, has begun (Longcor et al. 1996; L. Wong, L. Klionsky, N. Nangle, D. Mendinueto, V. Merriam, S. Wickert, J.D. Orias, E. Orias, and E. Hamilton, in prep.). Cloning biologically interesting mutant genes by complementation now seems within reach inTetrahymena, by exploiting the coassortment of mutant alleles with mapped DNA polymorphisms. The approach is to (1) identify the size of the ARP that carries the coassorting mutant gene, (2) size-select ARP DNA, (3) construct a minilibrary of inserts from this DNA, and (4) screen a relatively small number of Tetrahymena transformants for changed phenotype.
The Near Future
Immediate plans are to construct representative libraries of large, chromosome-specific MIC inserts, which would be available to any interested scientist. First priority will be given to the characterization of Cbs-containing cloned inserts. Insert ends, flanking the Cbs site, will be used to identify associated B–C3 RFLPs, each of which will then be mapped. The two ends of a given cloned insert should be closely linked in the MIC but should be on different MAC ARPs and thus should assort independently of one another in the MAC. Genetic and physical analyses of such ends should identify the sizes, order, and orientations of two ARPs mapping generally to adjacent segments in the MIC. When this work has been completed, we will have a map of MAC ARP contigs in a particular MIC chromosome, that is, a physical framework of each MIC chromosome.
Perspectives
Tetrahymena is one of very few eukaryotes with which the powers of molecular biology, conventional genetics, and microbial methodology can be synergistically combined. It is also the most distant relative of the plants, fungi, and vertebrates that has conventional meiosis, well-developed Mendelian genetic analysis, and an advanced genetic map. The long period of independent evolution from the vertebrates has endowed the ciliates with remarkable alternative uses of the fundamental eukaryotic biology, which enlighten and enrich our understanding of those mechanisms. Sequencing a ciliate genome is thus likely to make important contributions to our understanding of eukaryotic biology. Tetrahymena is a particularly important ciliate to sequence, given its biological versatility, the facility with which it can be cultivated under a variety of conditions, its experimental amenability, its utility as a research model system, and its potential for biotechnological applications.
With the availability of DNA polymorphism-based genetic maps of the MIC and MAC genomes, the groundwork has been laid for a concentrated effort to physically map and subsequently sequence the Tetrahymenagenome. International conferences held in the summer of 1997 (FASEB Conference in Ciliate Molecular Biology in Copper Mountain, CO, USA, and the Tenth International Congress of Protozoology in Sydney, Australia) collectively highlighted recent progress inTetrahymena genome mapping, DNA-mediated transformation, and biotechnological applications. Collaborative agreements made at these meetings are certain to speed up progress toward a matureTetrahymena genome project.
Acknowledgments
I thank Tim Lynch, Stephen Ng, Xueyu Shen, Eileen Hamilton, and Judy Orias for their indispensable help along the way in managing the Genome Mapping Project; the many talented and dedicated undergraduate students who identified and mapped most of the RAPDs; Sally L. Allen, Miu-Fun Chau, Eileen Hamilton, Tim Lynch, John Merriam, Virginia Merriam, Judy Orias, and Steve Wickert for critically reading the manuscript; the NIH National Research Resources Center, which continues to provide the main support for this effort (RO1-RR09231); and the UCSB undergraduate fellowship programs—Howard Hughes Medical Institute, California Alliance for Minority Participation, May Company, Genesis—which partially supported the research of many of the contributing undergraduate students. I regret that because of space limitations I have not been able to refer to the work of many talented investigators whose important efforts have exploited the useful features of ciliates.
Footnotes
-
↵1 E-MAIL ; FAX (805) 893-4724.
- Cold Spring Harbor Laboratory Press