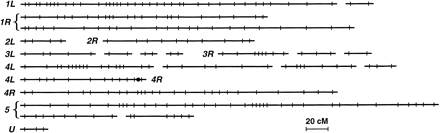
Maps of the MIC genome, by chromosome arm, based on ∼400 RAPD polymorphisms. (L and R) Left and right chromosome arms, except for chromosome 5, where arms cannot yet be distinguished. (U) Linkage group with uncertain chromosome assignment. Chromosomes throughout this review are numbered according to the recently revised scheme (J. Merriam, D. Cassidy-Hanley, and P.J. Bruns, pers. comm.). (•) The centromere region of chromosome 4, as RAPDs on its left and right map to 4L and 4R, respectively, by monosomic mapping. Other linkage groups within a chromosome arm remain to be oriented with respect to one another or to the centromere; this will be accomplished when more DNA polymorphisms are mapped or when immunocytological mapping inTetrahymena (D. Cassidy-Hanley and P.J. Bruns, pers. comm.) is used with the existing RAPDs. Most RAPDs have been mapped using only 32 meiotic segregants. Within a given linkage group, the order within a locus cluster and the orientation of clusters separated by large distances are still soft: Inversions often change the likelihood of the segregation data by <3 log units. Clusters of loci that failed to give recombinants with one another are represented by a single point. Loci unlinked to all others are not shown.











